Description:
My ultimate aim is to process ADNI data through the HCP pipeline, so I am attempting to use “import_bids”. For back ground me ADNI data has been converted using Clinca “adni-to-bids”
Looking at my bids folder versus the example on the BIDS support within QuNex page, I seem to be missing a “fmap” folder, which I have not seen or referred to before and could be the cause of my issue.
I am a bit confused about this warning, as I thought that path would start empty for the files to me moved/copied over too?
“===> WARNING:
No image files in bids folder!
There are no image files in the bids folder [/gpfs/home/tqh20wsu/scratch/qunex2/adni/sessions/ADNI002S1155/bids]
Please check your data!”
Finally, I don’t think I have my own parameters or mapping file, so I have used the demo quide ones, will that cause issues?
Thank you, Scott
Call:
qunex_container import_bids \
--sessionsfolder="${STUDY_FOLDER}/sessions" \
--inbox="${WORK_DIR}/bids" \
--action='copy' \
--archive='leave' \
--overwrite=no \
--bind="${WORK_DIR}:${WORK_DIR}" \
--container="${QUNEX_CONTAINER}"
Logs:
[tqh20wsu@c0004 ~]$ --> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXEXTENSIONS QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR PYLIBDIR FSLDIR FSLBINDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder EDDYCUDA USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT ANTSDIR CUDIMOT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.99.2
User: tqh20wsu
System: c0004.hpc.private
OS: RedHat Linux #1 SMP Thu Jan 25 16:17:31 UTC 2024
------------------------------------------------------------------------
██████\ ║ ██\ ██\
██ __██\ ║ ███\ ██ |
██ / ██ |██\ ██\ ║ ████\ ██ | ██████\ ██\ ██\
██ | ██ |██ | ██ | ║ ██ ██\██ |██ __██\\██\ ██ |
██ | ██ |██ | ██ | ║ ██ \████ |████████ |\████ /
██ ██\██ |██ | ██ | ║ ██ |\███ |██ ____|██ ██\
\██████ / \██████ | ║ ██ | \██ |\███████\██ /\██\
\___███\ \______/ ║ \__| \__| \_______\__/ \__|
\___| ║
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSES' which is a part of the QuNex Suite source code package:
https://gitlab.qunex.yale.edu/qunex/qunex/-/tree/master/LICENSES
---> Setting up Octave
........................ Running QuNex v0.99.2 ........................
--- Full QuNex call for command: import_bids
gmri import_bids --sessionsfolder="/gpfs/home/tqh20wsu/scratch/qunex2/adni/sessions" --inbox="/gpfs/home/tqh20wsu/scratch/qunex2/bids" --action="copy" --archive="leave" --overwrite="no"
---------------------------------------------------------
started running import_bids at 2024-04-24 16:50:14, track progress in /gpfs/home/tqh20wsu/scratch/qunex2/adni/processing/logs/comlogs/tmp_import_bids_2024-04-24_16.50.14.545950.log
call: gmri import_bids sessionsfolder="/gpfs/home/tqh20wsu/scratch/qunex2/adni/sessions" inbox="/gpfs/home/tqh20wsu/scratch/qunex2/bids" action="copy" archive="leave" overwrite="no"
-----------------------------------------
Running import_bids
==================
--> identifying files in /gpfs/home/tqh20wsu/scratch/qunex2/bids
--> Inbox type: bids_study
==> Paths:
bidsinfo -> /gpfs/home/tqh20wsu/scratch/qunex2/adni/info/bids
bidsinbox -> /gpfs/home/tqh20wsu/scratch/qunex2/adni/sessions/inbox/BIDS
bidsarchive -> /gpfs/home/tqh20wsu/scratch/qunex2/adni/sessions/archive/BIDS
--> mapping files to QuNex bids folders
--> processing BIDS info folder
--> creating bids session ADNI002S1155
--> creating bids session ADNI002S1155_M126
Running map_bids2nii for subject ADNI002S1155
=============================================
===> WARNING:
No image files in bids folder!
There are no image files in the bids folder [/gpfs/home/tqh20wsu/scratch/qunex2/adni/sessions/ADNI002S1155/bids]
Please check your data!
Running mapBIDS2behavior for subject ADNI002S1155
=================================================
--> created behavior subfolder
--> mapped: participants.tsv
Running map_bids2nii for subject ADNI002S1155, session M126
===========================================================
--> linked 1.nii.gz <-- sub-ADNI002S1155_ses-M126_T1w.nii.gz
--> linked 2.nii.gz <-- sub-ADNI002S1155_ses-M126_FLAIR.nii.gz
--> linked 3.nii.gz <-- sub-ADNI002S1155_ses-M126_task-rest_bold.nii.gz
Running mapBIDS2behavior for subject ADNI002S1155, session M126
===============================================================
--> created behavior subfolder
--> mapped: participants.tsv
Final report
============
subject ADNI002S1155 image mapping failed, behavioral files: 1 files mapped
subject ADNI002S1155, session M126 image mapping completed, behavioral files: 1 files mapped
===> ERROR in completing import_bids:
Some actions failed
Please check report!
-----------------------------------------
Finished at 2024-04-24 16:50:15
Hi,
The lack of fmap folder suggests that your imaging does not have this data, which means that the processing will be of lower quality. This is typical for older datasets. I am not 100% familiar with ADNI, so I cannot say if the lack of this is normal or if you did something wrong.
Mapping and parameter files come into play later on and do not influence the issue you are currently facing. You will have to amend them to suit your needs though. The parameter file defines the acquisition and processing parameters and will have to be tailored to the imaging data you are processing. Or else the results will be flawed/erroneous. The mapping data will have to be provided when mapping the data from QuNex folder structure to HCP folder structure, which is a step that follows what you are stuck on right now.
The error you are facing right now is:
There are no image files in the bids folder [/gpfs/home/tqh20wsu/scratch/qunex2/adni/sessions/ADNI002S1155/bids]
Please check your data!
Can you show us the contents of that directory? Could you also provide us with the structure (tree) of ${WORK_DIR}/bids? Thanks.
Best, Jure
1 Like
Hi Jure
Thank you for taking the time to get back to me.
I have liaised with Clinca whom I used to convert my ADNI data to BIDS format and they don’t currently use or output the field mapping.
Do you recommend any particular BIDS converter that outputs in the preferred structure for QuNex?
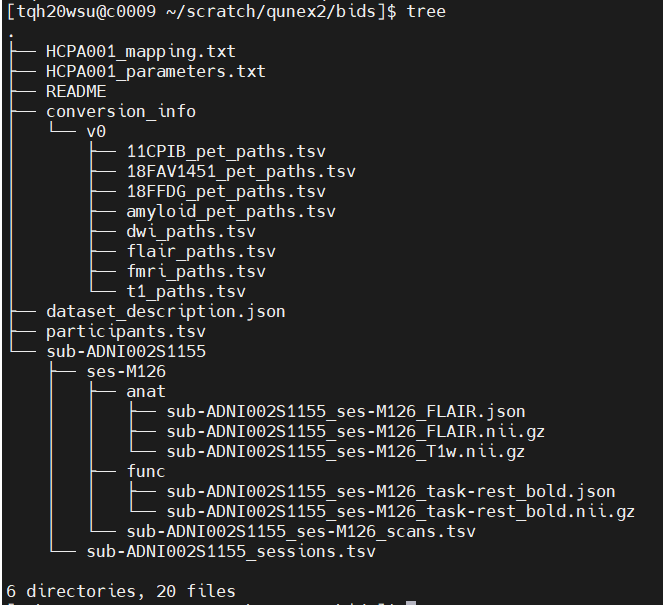
Below is the tree output for BIDS, which I think is correct accept the lack of fmap directory.
I noticed there is an import dicom method, could this possibly work with my ADNI data? I have put a tree of the raw data below too (I have removed most of the images to stay within the character limit):
[tqh20wsu@c0009 ~/Documents/Project/Pipelined/ADNI]$ tree
.
└── 002_S_1155
├── Accelerated_Sagittal_MPRAGE
│ └── 2017-04-24_13_21_32.0
│ └── I843510
│ ├── ADNI_002_S_1155_MR_Accelerated_Sagittal_MPRAGE__br_raw_20170425131856689_29_S558282_I843510.dcm
│ ├── ADNI_002_S_1155_MR_Accelerated_Sagittal_MPRAGE__br_raw_20170425131857032_169_S558282_I843510.dcm
│ ├── ADNI_002_S_1155_MR_Accelerated_Sagittal_MPRAGE__br_raw_20170425131857079_160_S558282_I843510.dcm
├── Axial_T2_STAR
│ └── 2017-04-24_13_21_32.0
│ └── I843511
│ ├── ADNI_002_S_1155_MR_Axial_T2_STAR__br_raw_20170425131857250_9_S558283_I843511.dcm
│ ├── ADNI_002_S_1155_MR_Axial_T2_STAR__br_raw_20170425131857609_16_S558283_I843511.dcm
│ ├── ADNI_002_S_1155_MR_Axial_T2_STAR__br_raw_20170425131857905_43_S558283_I843511.dcm
├── Axial_rsfMRI__Eyes_Open_
│ └── 2017-04-24_13_21_32.0
│ └── I843512
│ ├── ADNI_002_S_1155_MR_Axial_rsfMRI__Eyes_Open__br_raw_20170425131856501_29_S558284_I843512.dcm
│ ├── ADNI_002_S_1155_MR_Axial_rsfMRI__Eyes_Open__br_raw_20170425131856845_30_S558284_I843512.dcm
│ ├── ADNI_002_S_1155_MR_Axial_rsfMRI__Eyes_Open__br_raw_20170425131857172_11_S558284_I843512.dcm
├── Field_Mapping
│ └── 2017-04-24_13_21_32.0
│ ├── I843513
│ │ ├── ADNI_002_S_1155_MR_Field_Mapping__br_raw_20170425131857422_39_S558285_I843513.dcm
│ │ ├── ADNI_002_S_1155_MR_Field_Mapping__br_raw_20170425131858935_2_S558285_I843513.dcm
│ │ ├── ADNI_002_S_1155_MR_Field_Mapping__br_raw_20170425131859855_43_S558285_I843513.dcm
│ ├── I843520
│ │ ├── ADNI_002_S_1155_MR_Field_Mapping__br_raw_20170425131856579_31_S558292_I843520.dcm
│ │ ├── ADNI_002_S_1155_MR_Field_Mapping__br_raw_20170425131900620_19_S558292_I843520.dcm
│ │ ├── ADNI_002_S_1155_MR_Field_Mapping__br_raw_20170425131900916_51_S558292_I843520.dcm
│ └── I843521
│ ├── ADNI_002_S_1155_MR_Field_Mapping__br_raw_20170425131857500_7_S558285_I843521.dcm
│ ├── ADNI_002_S_1155_MR_Field_Mapping__br_raw_20170425131857656_12_S558285_I843521.dcm
│ ├── ADNI_002_S_1155_MR_Field_Mapping__br_raw_20170425131858124_42_S558285_I843521.dcm
└── Sagittal_3D_FLAIR
└── 2017-04-24_13_21_32.0
└── I843514
├── ADNI_002_S_1155_MR_Sagittal_3D_FLAIR__br_raw_20170425131856720_52_S558286_I843514.dcm
├── ADNI_002_S_1155_MR_Sagittal_3D_FLAIR__br_raw_20170425131857344_35_S558286_I843514.dcm
├── ADNI_002_S_1155_MR_Sagittal_3D_FLAIR__br_raw_20170425131857437_18_S558286_I843514.dcm
18 directories, 739 files
Thank you, Scott
Yes, import_dicom takes .dcm data and during the onboarding process converts it to .nii which is then used by all pipelines/analyses that follow. You can find an example of import_dicom on the Quick start page (QuNex quick start using a Docker container — QuNex documentation) under the Step-by-step execution section.
Let me know how the onboarding goes.
Hey Jure
I have attempted using import_dicom as per the instructions using the below calls
[tqh20wsu@c0004 ~]$ qunex_container create_study \
> --studyfolder="${STUDY_FOLDER}" \
> --container="${QUNEX_CONTAINER}"
[tqh20wsu@c0004 ~]$ qunex_container import_dicom \
> --sessionsfolder="${STUDY_FOLDER}/sessions" \
> --sessions="${SESSIONS}" \
> --masterinbox="${RAW_DATA}" \
> --archive="leave" \
> --container="${QUNEX_CONTAINER}"
[tqh20wsu@c0004 ~]$ qunex_container create_session_info \
> --sessionsfolder="${STUDY_FOLDER}/sessions" \
> --sessions="${SESSIONS}" \
> --mapping="${INPUT_MAPPING_FILE}" \
> --container="${QUNEX_CONTAINER}"
[tqh20wsu@c0004 ~]$ qunex_container setup_hcp \
> --sourcefolder="${STUDY_FOLDER}/sessions/${SESSIONS}" \
> --container="${QUNEX_CONTAINER}"
exported varriables:
QUNEX_CONTAINER=/gpfs/home/tqh20wsu/scratch/qunex2/qunex_suite-0.99.2d.sif
STUDY_NAME=quickstart_adni
STUDY_FOLDER=/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/
INPUT_BATCH_FILE=/gpfs/home/tqh20wsu/scratch/qunex2/data/HCPA001_parameters.txt
RAW_DATA=/gpfs/home/tqh20wsu/scratch/qunex2/ADNI
INPUT_MAPPING_FILE=/gpfs/home/tqh20wsu/scratch/qunex2/ADNI/HCPA001_mapping.txt
PATH=/gpfs/software/ada/tree/0.1:/gpfs/home/tqh20wsu/scratch/qunex2:/gpfs/software/ada/singularity/3.6.0/libexec/singularity/bin:/gpfs/software/ada/singularity/3.6.0/bin:/gpfs/software/ada/golang/1.13.4/go/bin:/gpfs/software/ada/python/anaconda/2023.07-2/bin:/gpfs/home/tqh20wsu/miniconda3/bin:/gpfs/home/tqh20wsu/bin:/usr/local/UEA/bin:/usr/local/bin:/usr/bin:/usr/local/sbin:/usr/sbin:/usr/lpp/mmfs/bin
RUNTURNKEY_TYPE=local
SESSIONS=002_S_1155
WORK_DIR=/gpfs/home/tqh20wsu/scratch/qunex2
HOME=/gpfs/home/tqh20wsu
Below is a tree of what I have within the QuNex folder so far, which also includes the ADNI folder I mentioned earlier:
├── quickstart_adni
│ ├── analysis
│ │ └── scripts
│ ├── info
│ │ ├── bids
│ │ ├── demographics
│ │ ├── hcpls
│ │ ├── stimuli
│ │ └── tasks
│ ├── processing
│ │ ├── lists
│ │ ├── logs
│ │ │ ├── batchlogs
│ │ │ │ ├── qunex_container_command_2024-04-25_21.40.11.410814.sh
│ │ │ │ └── qunex_container_command_2024-04-25_21.41.14.028653.sh
│ │ │ ├── comlogs
│ │ │ │ ├── done_create_session_info_2024-04-25_21.41.17.399831.log
│ │ │ │ ├── done_create_study_2024-04-25_21.39.59.040975.log
│ │ │ │ ├── done_import_dicom_2024-04-25_21.40.14.779195.log
│ │ │ │ └── error_setup_hcp_2024-04-25_21.41.33.293863.log
│ │ │ ├── runchecks
│ │ │ └── runlogs
│ │ │ ├── Log-create_session_info_2024-04-25_21.41.17.399831.log
│ │ │ ├── Log-create_study_2024-04-25_21.39.59.040975.log
│ │ │ ├── Log-import_dicom_2024-04-25_21.40.14.779195.log
│ │ │ └── Log-setup_hcp_2024-04-25_21.41.33.293863.log
│ │ ├── scenes
│ │ │ └── QC
│ │ │ ├── BOLD
│ │ │ ├── DWI
│ │ │ ├── T1w
│ │ │ ├── T2w
│ │ │ └── myelin
│ │ └── scripts
│ └── sessions
│ ├── 002_S_1155
│ │ ├── dicom
│ │ │ ├── 130.tar.gz
│ │ │ ├── 140.tar.gz
│ │ │ ├── 150.tar.gz
│ │ │ ├── 20.tar.gz
│ │ │ ├── 30.tar.gz
│ │ │ ├── 40.tar.gz
│ │ │ ├── DICOM-Report.txt
│ │ │ ├── dcm2niix_130.log
│ │ │ ├── dcm2niix_140.log
│ │ │ ├── dcm2niix_150.log
│ │ │ ├── dcm2niix_20.log
│ │ │ ├── dcm2niix_30.log
│ │ │ └── dcm2niix_40.log
│ │ ├── hcp
│ │ │ └── 002_S_1155
│ │ │ └── unprocessed
│ │ ├── inbox
│ │ │ └── 1
│ │ ├── nii
│ │ │ ├── 131.json
│ │ │ ├── 131.nii.gz
│ │ │ ├── 132.json
│ │ │ ├── 132.nii.gz
│ │ │ ├── 141.json
│ │ │ ├── 141.nii.gz
│ │ │ ├── 151.json
│ │ │ ├── 151.nii.gz
│ │ │ ├── 21.json
│ │ │ ├── 21.nii.gz
│ │ │ ├── 31.json
│ │ │ ├── 31.nii.gz
│ │ │ ├── 41.json
│ │ │ └── 41.nii.gz
│ │ ├── session.txt
│ │ └── session_hcp.txt
│ ├── QC
│ ├── archive
│ │ ├── BIDS
│ │ ├── EEG
│ │ ├── HCPLS
│ │ ├── MR
│ │ └── behavior
│ ├── inbox
│ │ ├── BIDS
│ │ ├── EEG
│ │ ├── HCPLS
│ │ ├── MR
│ │ ├── behavior
│ │ ├── concs
│ │ └── events
│ └── specs
│ └── parameters_example.txt
├── qunex_container
└── qunex_suite-0.99.2d.sif
error_setup_hcp_2024-04-25_21.41.33.293863.log (1.1 KB)
done_import_dicom_2024-04-25_21.40.14.779195.log (112.4 KB)
done_create_study_2024-04-25_21.39.59.040975.log (3.8 KB)
done_create_session_info_2024-04-25_21.41.17.399831.log (757 Bytes)
Good progress :). The data got onboarded successfully.
What you will need to do is amend the mapping file. The problem is that we are usually unable to extract all the metadata about the images that we need reliably from dicoms, so we need to provide some information to QuNex manually via the mapping file. See Preparing a study-level mapping file for QuNex workflows — QuNex documentation for additional details. To help you out, I would need 2 things:
- The
session.txt file found at <qx_study>/sessions/002_S_1155/session.txt.
- The mapping file you are using
INPUT_MAPPING_FILE=/gpfs/home/tqh20wsu/scratch/qunex2/ADNI/HCPA001_mapping.txt. I guess this is the one from the quick start. Mapping file is study/acquisition specific and needs to be tweaked for every study. If the acquisition protocol is not consistent across sessions in a study, you need to use a different mapping for sessions that difer.
Once we sort that out, you will rerun create_session_info and setup_hcp and after that you will be set to proceed with HCP pipelines. So you are nearly there.
Best, Jure
Hi Jure,
Sorry for the bleated reply, I had a busy Friday so couldn’t get back to you sooner.
Thank you once again for your continued help and support, it is greatly appreciated.
I have attached the files requested, including my attempts at updating them.
session_org.txt (702 Bytes)
session.txt (759 Bytes)
HCPA001_mapping.txt (508 Bytes)
adni_mapping.txt (90 Bytes)
I re-ran create_session_info without any issue and attempted setup_hcp again but I got a new error (log also attached)
error_setup_hcp_2024-04-28_19.46.22.625068.log (1.1 KB)
---> Base folder /gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/sessions/002_S_1155/hcp/002_S_1155/unprocessed already exist! Adding any new files specified!
... skipping 151 [unknown sequence label, please check]
... skipping 141 [unknown sequence label, please check]
... skipping 132 [unknown sequence label, please check]
... skipping 131 [unknown sequence label, please check]
... skipping 41 [unknown sequence label, please check]
... skipping 31 [unknown sequence label, please check]
... skipping 21 [unknown sequence label, please check]
===> ERROR in completing setup_hcp:
No files mapped
No files were found to be mapped to the hcp folder [/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/sessions/002_S_1155]!
Please check your data!
Initially I only updated the #_mappings.txt file so went back and amended sessions.txt as per an old forum post but that did not seem to make a noticeable difference.
My ADNI data for this one test subject includes two options for T2w, will including/pairing both cause any issues? Should I pick just one?
Finally I am unsure if the field maps I have are Gradient, Magnitude or Phase so I excluded any association within the mapping and session files.
Thank you again, Scott
Hi, manual edits to the session.txt file should not be needed, those are required when you are doing some unconventional processing. You are not doing that so all should be good here.
The mapping file you attached looks good. How this works is that the session.txt file is created when data is onboarded. That file is populated with information/meta-data that can be reliably extracted from imaging data. Once you run create_session_info a file called session_hcp.txt is created that includes the extra information that is required for HCP processing and once you have that, you can run setup_hcp to prepare the data for HCP processing.
Please do this:
- Remove
session_hcp.txt in the folder for the session.
- Restore original
session.txt.
- Rerun
create_session_info by using the adni_mapping.txt you provided.
- If
create_session_info finishes successfully, inspect session_hcp.txt and upload it here so I can see if it worked.
If all is well, we will then proceed with setup_hcp and HCP processing.
Best, Jure
Good morning, Jure
Looks like it worked, will give setup_hcp a go
session_hcp.txt (794 Bytes)
Thank you, Scott
Looks like it worked
done_setup_hcp_2024-04-29_11.04.16.383783.log (1.1 KB)
I assume I work my way through these now?
create_batch,hcp_pre_freesurfer,hcp_freesurfer,hcp_post_freesurfer,hcp_fmri_volume,hcp_fmri_surface
Or should I go back to the turnkey approach?
Kind regards, Scott
edit to add:
create_batch looked complicated with the parameters.txt file needing to be created.
Going to try turnkey with the rest of the jobs
edit2:
Seems I need a parameters.txt file, will wait for your reply
Glad it worked. I would continue with a step-by-step approach as it gives you more control than the run_turnkey approach. Also, run_turnkey has some limitations and issues and we are coding a major rework of the whole funcitonality.
I am pasting the next couple of commands below. For the parameters file, you can use the one that is shipped with quickstart. However, you might have to open it and tune some parameters based on the ADNI acquisition protocol, that one is for HCP style acquisition with a Siemens 3T scanner. So to get optimal results maybe you will have to tweak some things. I am not familiar with teh ADNI acquisition protocol, so I cannot really help you out much here.
Next commands:
STUDY_FOLDER="/data/qx_studies/quickstart" # <--- tweak this!
SESSIONS_FOLDER="${STUDY_FOLDER}/sessions"
BATCHFILE="${STUDY_FOLDER}/processing/batch.txt"
CONTAINER="/qunex_containers/qunex_suite-0.99.2d.sif" # <--- tweak this!
qunex_container create_batch \
--sessionsfolder="${SESSIONS_FOLDER}" \
--sessions="HCPA001" \ # <--- tweak this!
--paramfile="/specs/HCP_parameters.txt" \ # <--- tweak this!
--targetfile="${BATCHFILE}" \
--container="${CONTAINER}"
qunex_container hcp_pre_freesurfer \
--sessionsfolder="${SESSIONS_FOLDER}" \
--batchfile="${BATCHFILE}" \
--container="${CONTAINER}"
qunex_container hcp_freesurfer \
--sessionsfolder="${SESSIONS_FOLDER}" \
--batchfile="${BATCHFILE}" \
--container="${CONTAINER}"
qunex_container hcp_post_freesurfer \
--sessionsfolder="${SESSIONS_FOLDER}" \
--batchfile="${BATCHFILE}" \
--container="${CONTAINER}"
Hi Jure
I have an issue with hcp_pre_freesurfer and my thought is its because I have 2x t2 images.
Should I go back and update my sessions.txt and run the steps again?
Thank you, Scott
error_hcp_pre_freesurfer_002_S_1155_2024-04-29_16.19.29.293038.log (5.0 KB)
I tried again, removing one of the T2w pairing from the mappings.txt but it came back with the same error
error_hcp_pre_freesurfer_002_S_1155_2024-04-29_18.09.24.155239.log (4.6 KB)
Hi,
No, this is not because of a double T2w. You can see in the log that T2w images are correctly assigned. The error is this:
Mon Apr 29 16:19:29 BST 2024:PreFreeSurferPipeline.sh: ERROR: missing the
following mandatory parameter(s): --seechospacing --seunwarpdir --t1samplespacing
--t2samplespacing --unwarpdir
Could you upload your batchfile. It seems like the parameters are not set in there (they should be in the quickstart parameters file). In any case they should be set for your dataset and not just copied from somewhere. You will get sub-optimal/erroneous results if these are not set correctly.
Best, Jure
Hi Jure
Here is the file as requested
I dont think I copied/move this from anywhere it was just created
Thank you, Scott
batch.txt (44.6 KB)
Hi,
If you open the attached batch file, you can see that all of the parameters are commented (the lines start with # so they do not get propagated into commands). In the batch file, you first have parameters that get propagated to QuNex commands (to assure consistency across command runs), followed by session info.
IT seems like here:
qunex_container create_batch \
--sessionsfolder="${SESSIONS_FOLDER}" \
--sessions="002_S_1155"
--paramfile="/specs/HCP_parameters.txt" \ # <--- tweak this!
--targetfile="${BATCHFILE}" \
--container="${CONTAINER}"
You did not correctly tweak the location of the param file. As a starting point, you can use the Quick Start one (http://jd.mblab.si/qunex/HCPA001_parameters.txt). But like I said, to get optimal or even meaningful results, you probably need to set some parameters in accordance with the ADNI acquisition protocol and device.
Best, Jure
Hi Jure
So I reran:
[tqh20wsu@c0008 ~]$ module load singularity/3.6.0
[tqh20wsu@c0008 ~]$ PATH=${HOME}/scratch/qunex2:${PATH}
[tqh20wsu@c0008 ~]$ export STUDY_NAME="quickstart_adni"
[tqh20wsu@c0008 ~]$ export WORK_DIR="${HOME}/scratch/qunex2"
[tqh20wsu@c0008 ~]$ export STUDY_FOLDER="${WORK_DIR}/${STUDY_NAME}"
[tqh20wsu@c0008 ~]$ export QUNEX_CONTAINER=${WORK_DIR}/qunex_suite-0.99.2d.sif
[tqh20wsu@c0008 ~]$ export RAW_DATA="${WORK_DIR}/ADNI"
[tqh20wsu@c0008 ~]$ export INPUT_BATCH_FILE="${RAW_DATA}/HCPA001_parameters.txt"
[tqh20wsu@c0008 ~]$ export INPUT_MAPPING_FILE="${RAW_DATA}/adni_mapping.txt"
[tqh20wsu@c0008 ~]$ export SESSIONS="002_S_1155"
[tqh20wsu@c0008 ~]$ export RUNTURNKEY_TYPE="local"
[tqh20wsu@c0008 ~]$
[tqh20wsu@c0008 ~]$ export SESSIONS_FOLDER="${STUDY_FOLDER}/sessions"
[tqh20wsu@c0008 ~]$ export BATCHFILE="${STUDY_FOLDER}/processing/batch.txt"
[tqh20wsu@c0008 ~]$ export CONTAINER="${QUNEX_CONTAINER}"
[tqh20wsu@c0008 ~]$ export PARAMS="${SESSIONS_FOLDER}/specs/parameters.txt"
[tqh20wsu@c0008 ~]$ qunex_container create_batch \
> --sessionsfolder="${SESSIONS_FOLDER}" \
> --sessions="${SESSIONS}" \
> --paramfile="${PARAMS}" \
> --targetfile="${BATCHFILE}" \
> --container="${CONTAINER}" \
> --overwrite=”yes”
[tqh20wsu@c0008 ~]$ qunex_container hcp_pre_freesurfer \
> --sessionsfolder="${SESSIONS_FOLDER}" \
> --batchfile="${BATCHFILE}" \
> --container="${CONTAINER}"
Here is the new batch.txt file
batch.txt (5.4 KB)
And got the following output:
--> QuNex will run the command over 1 sessions. It will utilize:
Maximum sessions run in parallel for a job: 1.
Maximum elements run in parallel for a session: 1.
Up to 1 processes will be utilized for a job.
Job #1 will run sessions: 002_S_1155
[tqh20wsu@c0008 ~]$ --> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXEXTENSIONS QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR PYLIBDIR FSLDIR FSLBINDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder EDDYCUDA USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT ANTSDIR CUDIMOT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.99.2
User: tqh20wsu
System: c0008.hpc.private
OS: RedHat Linux #1 SMP Thu Jan 25 16:17:31 UTC 2024
------------------------------------------------------------------------
██████\ ║ ██\ ██\
██ __██\ ║ ███\ ██ |
██ / ██ |██\ ██\ ║ ████\ ██ | ██████\ ██\ ██\
██ | ██ |██ | ██ | ║ ██ ██\██ |██ __██\\██\ ██ |
██ | ██ |██ | ██ | ║ ██ \████ |████████ |\████ /
██ ██\██ |██ | ██ | ║ ██ |\███ |██ ____|██ ██\
\██████ / \██████ | ║ ██ | \██ |\███████\██ /\██\
\___███\ \______/ ║ \__| \__| \_______\__/ \__|
\___| ║
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSES' which is a part of the QuNex Suite source code package:
https://gitlab.qunex.yale.edu/qunex/qunex/-/tree/master/LICENSES
---> Setting up Octave
........................ Running QuNex v0.99.2 ........................
--- Full QuNex call for command: hcp_pre_freesurfer
gmri hcp_pre_freesurfer --sessionsfolder="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/sessions" --batchfile="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/processing/batch.txt" --sessions="002_S_1155"
---------------------------------------------------------
WARNING: Use of parameters with changed name(s)!
The following parameters have new names:
... TR is now tr!
Please correct the listed parameter names in command line or batch file!
WARNING: Parameter qx_cifti_tail was not specified. Its value was imputed from parameter hcp_cifti_tail and set to '_Atlas'!
WARNING: Parameter qx_nifti_tail was not specified. Its value was imputed from parameter hcp_nifti_tail and set to ''!
WARNING: Parameter cifti_tail was not specified. Its value was imputed from parameter qx_cifti_tail and set to '_Atlas'!
WARNING: Parameter nifti_tail was not specified. Its value was imputed from parameter qx_nifti_tail and set to ''!
# Generated by QuNex 0.99.2 on 2024-05-01_16.06.15.970383#
=================================================================
gmri hcp_pre_freesurfer \
--sessionsfolder="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/sessions" \
--sessions="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/processing/batch.txt" \
--sessionids="002_S_1155" \
=================================================================
Starting multiprocessing sessions in /gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/processing/batch.txt with a pool of 1 concurrent processes
---- Running local
Starting processing of sessions 002_S_1155 at Wednesday, 01. May 2024 16:06:15
------------------------------------------------------------
Session id: 002_S_1155
[started on Wednesday, 01. May 2024 16:06:15]
Running HCP PreFreeSurfer Pipeline [HCPStyleData] ...
---> T1w image file present.
---> T2w image file present.
---> ERROR: Could not find folder with files for TOPUP processing of session 002_S_1155.
ERROR: Unknown error occured:
...................................
Traceback (most recent call last):
File "/opt/qunex/python/qx_utilities/hcp/process_hcp.py", line 854, in hcp_pre_freesurfer
tufolder = glob.glob(
^^^^^^^^^^
IndexError: list index out of range
...................................
HCP PreFS completed on Wednesday, 01. May 2024 16:06:15
------------------------------------------------------------
===> Final report for command hcp_pre_freesurfer
... 002_S_1155 ---> PreFS failed
===> Not all tasks completed fully!
These are the parameters I was talking about. I am not familiar with ADNI acquisition device or protocol so I cannot really help you set all of this optimally. Based on the session files that I saw, I believe you should either set the line 28 in batch.txt to:
...
_hcp_avgrdcmethod : NONE
...
or add the parameter below to your hcp_pre_freesurfer call:
...
--hcp_avgrdcmethod="NONE"
...
Best, Jure
Hi Jure
Looking at the docs this setting relats to corrects using the field maps, correct?
Unfortunately, ADNI doesn’t obviously say if their field maps are Gradient, Magnitude or Phase. I asked on their forum 4 days ago but noone has responded.
Looking at the data I do have for one of the field maps, I can at least tell it is Siemens, but I will focus on optimal outputs once I have the pipeline work and am process more data.
I will set to None as per your suggestion and report back.
Thank you, Scott
Hi Jure
It appears to be running ok, I will leave it for a bit and see how it gets on.
Thank you again for your continued assistance, Scott
[tqh20wsu@c0004 ~]$ qunex_container hcp_pre_freesurfer \
> --sessionsfolder="${SESSIONS_FOLDER}" \
> --batchfile="${BATCHFILE}" \
> --container="${CONTAINER}"
--> QuNex will run the command over 1 sessions. It will utilize:
Maximum sessions run in parallel for a job: 1.
Maximum elements run in parallel for a session: 1.
Up to 1 processes will be utilized for a job.
Job #1 will run sessions: 002_S_1155
[tqh20wsu@c0004 ~]$ --> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXEXTENSIONS QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR PYLIBDIR FSLDIR FSLBINDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder EDDYCUDA USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT ANTSDIR CUDIMOT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.99.2
User: tqh20wsu
System: c0004.hpc.private
OS: RedHat Linux #1 SMP Thu Jan 25 16:17:31 UTC 2024
------------------------------------------------------------------------
██████\ ║ ██\ ██\
██ __██\ ║ ███\ ██ |
██ / ██ |██\ ██\ ║ ████\ ██ | ██████\ ██\ ██\
██ | ██ |██ | ██ | ║ ██ ██\██ |██ __██\\██\ ██ |
██ | ██ |██ | ██ | ║ ██ \████ |████████ |\████ /
██ ██\██ |██ | ██ | ║ ██ |\███ |██ ____|██ ██\
\██████ / \██████ | ║ ██ | \██ |\███████\██ /\██\
\___███\ \______/ ║ \__| \__| \_______\__/ \__|
\___| ║
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSES' which is a part of the QuNex Suite source code package:
https://gitlab.qunex.yale.edu/qunex/qunex/-/tree/master/LICENSES
---> Setting up Octave
........................ Running QuNex v0.99.2 ........................
--- Full QuNex call for command: hcp_pre_freesurfer
gmri hcp_pre_freesurfer --sessionsfolder="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/sessions" --batchfile="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/processing/batch.txt" --sessions="002_S_1155"
---------------------------------------------------------
WARNING: Use of parameters with changed name(s)!
The following parameters have new names:
... TR is now tr!
Please correct the listed parameter names in command line or batch file!
WARNING: Parameter qx_cifti_tail was not specified. Its value was imputed from parameter hcp_cifti_tail and set to '_Atlas'!
WARNING: Parameter qx_nifti_tail was not specified. Its value was imputed from parameter hcp_nifti_tail and set to ''!
WARNING: Parameter cifti_tail was not specified. Its value was imputed from parameter qx_cifti_tail and set to '_Atlas'!
WARNING: Parameter nifti_tail was not specified. Its value was imputed from parameter qx_nifti_tail and set to ''!
# Generated by QuNex 0.99.2 on 2024-05-02_11.21.34.367143#
=================================================================
gmri hcp_pre_freesurfer \
--sessionsfolder="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/sessions" \
--sessions="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/processing/batch.txt" \
--sessionids="002_S_1155" \
=================================================================
Starting multiprocessing sessions in /gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/processing/batch.txt with a pool of 1 concurrent processes
---- Running local
Starting processing of sessions 002_S_1155 at Thursday, 02. May 2024 11:21:34
Running external command: /opt/HCP/HCPpipelines/PreFreeSurfer/PreFreeSurferPipeline.sh --path="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/sessions/002_S_1155/hcp" --subject="002_S_1155" --t1="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/sessions/002_S_1155/hcp/002_S_1155/unprocessed/T1w/002_S_1155_T1w_MPR1.nii.gz" --t2="/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/sessions/002_S_1155/hcp/002_S_1155/unprocessed/T2w/002_S_1155_T2w_SPC1.nii.gz" --t1template="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_0.7mm.nii.gz" --t1templatebrain="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_0.7mm_brain.nii.gz" --t1template2mm="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm.nii.gz" --t2template="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_0.7mm.nii.gz" --t2templatebrain="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_0.7mm_brain.nii.gz" --t2template2mm="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_2mm.nii.gz" --templatemask="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_0.7mm_brain_mask.nii.gz" --template2mmmask="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm_brain_mask_dil.nii.gz" --brainsize="150" --fnirtconfig="/opt/HCP/HCPpipelines/global/config/T1_2_MNI152_2mm.cnf" --seechospacing="0.000580009" --seunwarpdir="y" --t1samplespacing="0.0000071" --t2samplespacing="0.0000021" --unwarpdir="z" --gdcoeffs="NONE" --avgrdcmethod="NONE" --processing-mode="HCPStyleData"
You can follow command's progress in:
/gpfs/home/tqh20wsu/scratch/qunex2/quickstart_adni/processing/logs/comlogs/tmp_hcp_pre_freesurfer_002_S_1155_2024-05-02_11.21.34.389335.log
------------------------------------------------------------