Hi Jure!
I have just started using QuNex containers to process my data and due to the amount of data I have to process, I would like to be able to run QuNex on a cluster (hpc) using singularity containers. but I am still not able to run QuNex correctly even after I have tried various ways of running it. i really need your help!
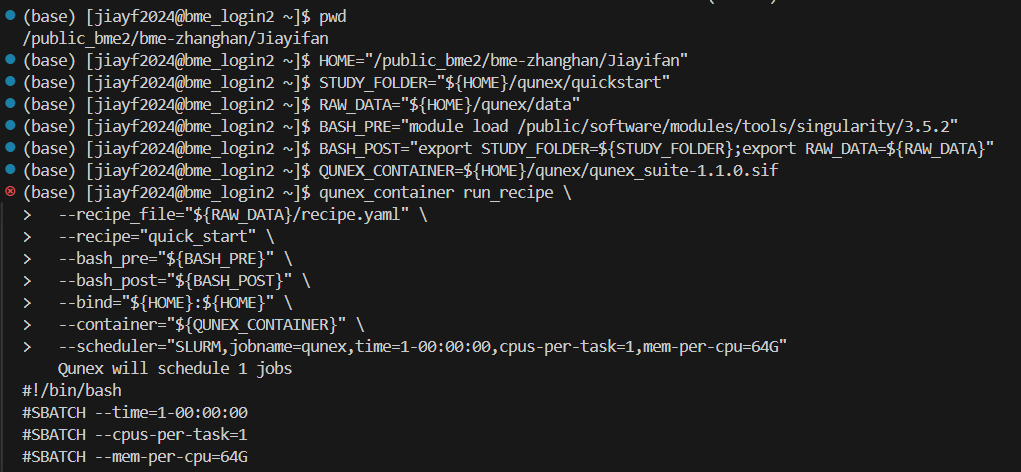
①I have tried directly at the login node, executing the following command:
STUDY_FOLDER="${HOME}/qunex/quickstart"
RAW_DATA="${HOME}/qunex/data"
BASH_POST="export STUDY_FOLDER=${STUDY_FOLDER};export RAW_DATA=${RAW_DATA}"
QUNEX_CONTAINER=${HOME}/containers/qunex_suite-1.1.0.sif
qunex_container run_recipe \
--recipe_file=“${RAW_DATA}/recipe.yaml” \
--recipe=“quick_start” \
--bash_post=“${BASH_POST}” \
--container="${QUNEX_CONTAINER}” \
---bind=“${HOME}:${HOME}” \
--container="${QUNEX_CONTAINER}” \
---scheduler="SLURM,jobname=qunex,time=1-00:00:00,cpus-per-task=1,mem-per-cpu=64G”
However, there is no output result, and the terminal interface is as follows:
(base) [jiayf2024@bme_login2 ~]$ qunex_container run_recipe \
> --recipe_file="${RAW_DATA}/recipe.yaml" \
> --recipe="quick_start" \
> --bash_post="${BASH_POST}" \
> --container="${QUNEX_CONTAINER}" \
> --bind="${HOME}:${HOME}" \
> --container="${QUNEX_CONTAINER}" \
> --scheduler="SLURM,jobname=qunex,time=1-00:00:00,cpus-per-task=1,mem-per-cpu=64G"
Qunex will schedule 1 jobs
#!/bin/bash
#SBATCH --time=1-00:00:00
#SBATCH --cpus-per-task=1
#SBATCH --mem-per-cpu=64G
②I also tried writing ‘qunex_container’ related commands in a newly created ‘.job’ file, but after running for more than 10 seconds, the job ends very quickly without any error reporting or any processing results.The contents of the ‘.job’ job are as follows:
#!/bin/bash
#SBATCH -J qunex
#SBATCH -p bme_cpu
#SBATCH --cpus-per-task=4
#SBATCH --mail-type=fail
#SBATCH -o job%j.out
#SBATCH -e job%j.err
#SBATCH -N 1
hostname
date
module load /public/software/modules/tools/singularity/3.5.2
chmod -R 777 /public_bme2/bme-zhanghan/Jiayifan/qunex
singularity run -e -B /public_bme2/bme-zhanghan/Jiayifan/qunex/data:/data /public_bme2/bme-zhanghan/Jiayifan/qunex/qunex_suite-1.1.0.sif
HOME="/public_bme2/bme-zhanghan/Jiayifan"
STUDY_FOLDER="/public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart"
RAW_DATA="/public_bme2/bme-zhanghan/Jiayifan/qunex/data"
BASH_POST="export STUDY_FOLDER=${STUDY_FOLDER};export RAW_DATA=${RAW_DATA}"
QUNEX_CONTAINER="/public_bme2/bme-zhanghan/Jiayifan/qunex/qunex_suite-1.1.0.sif"
qunex_container run_recipe \
--recipe_file="${RAW_DATA}/recipe.yaml" \
--recipe="quick_start" \
--bash_post="${BASH_POST}" \
--container="${QUNEX_CONTAINER}" \
--bind="${HOME}:${HOME}" \
--container="${QUNEX_CONTAINER}" \
--scheduler="SLURM,jobname=qunex,time=1-00:00:00,cpus-per-task=1,mem-per-cpu=64G"
③I have also tried executing the command after the login node enters the singularity container, at which point the command can be executed with the following output:
(base) [jiayf2024@bme_login2 ~]$ module load /public/software/modules/tools/singularity/3.5.2
(base) [jiayf2024@bme_login2 qunex]$ singularity run -e -B /public_bme2/bme-zhanghan/Jiayifan/qunex/data:/data /public_bme2/bme-zhanghan/Jiayifan/qunex/qunex_suite-1.1.0.sif
$ source /opt/qunex/env/qunex_environment.sh
---> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXEXTENSIONS QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR PYLIBDIR FSLDIR FSLBINDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_FS_CUSTOM HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder EDDYCUDA USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT ANTSDIR CUDIMOT
========================================================================
Generated by QuNex
------------------------------------------------------------------------
Version: 1.1.0 [QIO]
User: jiayf2024
System: bme_login2
OS: Debian Linux #1 SMP Mon Dec 7 11:30:56 UTC 2020
------------------------------------------------------------------------
██████\ ║ ██\ ██\
██ __██\ ║ ███\ ██ |
██ / ██ |██\ ██\ ║ ████\ ██ | ██████\ ██\ ██\
██ | ██ |██ | ██ | ║ ██ ██\██ |██ __██\\██\ ██ |
██ | ██ |██ | ██ | ║ ██ \████ |████████ |\████ /
██ ██\██ |██ | ██ | ║ ██ |\███ |██ ____|██ ██\
\██████ / \██████ | ║ ██ | \██ |\███████\██ /\██\
\___███\ \______/ ║ \__| \__| \_______\__/ \__|
\___| ║
DEVELOPED & MAINTAINED BY:
Mind & Brain Lab, University of Ljubljana
Cho Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
QuNex LICENSES which can be found in the LICENSES folder of the QuNex
repository or at https://qunex.yale.edu/qunex-registration
========================================================================
---> Setting up Octave
(qunex) [bme_login2 qunex]$ STUDY_FOLDER="/public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart"
(qunex) [bme_login2 qunex]$ QUNEX_CONTAINER="/public_bme2/bme-zhanghan/Jiayifan/qunex/qunex_suite-1.1.0.sif"
(qunex) [bme_login2 qunex]$ qunex_container create_study \
--studyfolder="${STUDY_FOLDER}" \
--container="${QUNEX_CONTAINER}"
/bin/sh: 1: singularity: not found
(qunex) [bme_login2 qunex]$ qunex create_study --studyfolder="${STUDY_FOLDER}" --container="${QUNEX_CONTAINER}"
.......................... Running QuNex v1.1.0 [QIO] ..........................
--- Full QuNex call for command: create_study
qunex create_study --studyfolder="/public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart" --container="/public_bme2/bme-zhanghan/Jiayifan/qunex/qunex_suite-1.1.0.sif"
---------------------------------------------------------
ERROR: Extra argument container is not valid! Please check your command!
ERROR in running create_study at gmri:
Invalid arguments
Provided unknown arguments: [container]!
The call received was:
(please note that when run through scheduler, all possible parameters,
even non relevant ones are passed)
qunex create_study \
--studyfolder=/public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart \
--container=/public_bme2/bme-zhanghan/Jiayifan/qunex/qunex_suite-1.1.0.sif
Please run `qunex create_study --help` to get help for the failed command.
(qunex) [bme_login2 qunex]$ qunex create_study --studyfolder="/public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart"
.......................... Running QuNex v1.1.0 [QIO] ..........................
--- Full QuNex call for command: create_study
qunex create_study --studyfolder="/public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart"
---------------------------------------------------------
started running create_study at 2025-03-09 13:45:50, track progress in /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/logs/comlogs/tmp_create_study_2025-03-09_13.45.50.774791.log
call: gmri create_study studyfolder="/public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart"
-----------------------------------------
Running create_study
===================
Creating study folder structure:
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/analysis
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/analysis/scripts
... folder exists: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing
... folder exists: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/logs
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/logs/batchlogs
... folder exists: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/logs/comlogs
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/logs/runlogs
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/lists
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/scripts
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/scenes
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/scenes/QC
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/scenes/QC/T1w
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/scenes/QC/T2w
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/scenes/QC/myelin
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/scenes/QC/BOLD
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/scenes/QC/DWI
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/info
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/info/demographics
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/info/tasks
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/info/stimuli
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/info/bids
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/info/hcpls
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/inbox
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/inbox/MR
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/inbox/EEG
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/inbox/BIDS
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/inbox/HCPLS
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/inbox/behavior
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/inbox/concs
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/inbox/events
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/archive
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/archive/MR
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/archive/EEG
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/archive/BIDS
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/archive/HCPLS
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/archive/behavior
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/specs
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/sessions/QC
... created: /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/subjects
Preparing template files:
... created parameters_example.txt file
... created .qunexstudy file
-----------------------------------------
Finished at 2025-03-09 13:45:50
---> Successful completion of task
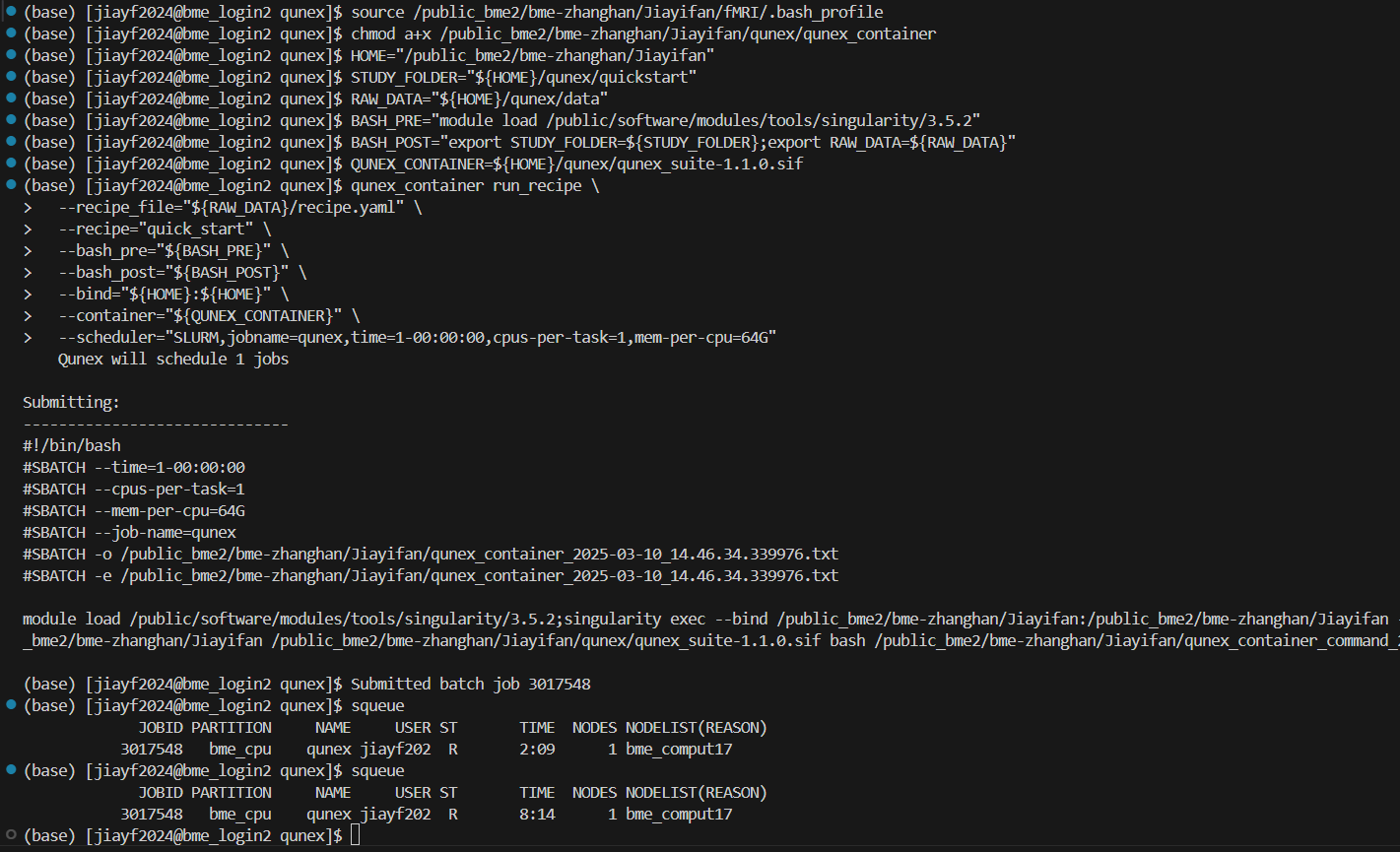
The third method, while it can execute some simple commands, I’m not sure it’s running on the login node, and although I subsequently added the ‘-scheduler’ parameter, I feel that the addition of that parameter rather prevents the commands from running for the following reasons:
(qunex) [bme_login2 qunex]$ qunex create_study \
> --studyfolder="${STUDY_FOLDER}" \
> --scheduler="SLURM,jobname=qunex,time=1-00:00:00,cpus-per-task=1,mem-per-cpu=64G"
.......................... Running QuNex v1.1.0 [QIO] ..........................
--- Full QuNex call for command: create_study
qunex create_study --studyfolder="/public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart" --scheduler="SLURM,jobname=qunex,time=1-00:00:00,cpus-per-task=1,mem-per-cpu=64G"
---------------------------------------------------------
---> Running scheduler for command create_study
---> submitting create_study
gmri create_study --studyfolder="/public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart"
Submitting:
------------------------------
#!/bin/bash
#SBATCH --time=1-00:00:00
#SBATCH --cpus-per-task=1
#SBATCH --mem-per-cpu=64G
#SBATCH --job-name=qunex
#SBATCH -o /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/logs/SLURM_create_study.2025-03-09_13.50.40.712483.log
#SBATCH -e /public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart/processing/logs/SLURM_create_study.2025-03-09_13.50.40.712483.log
gmri create_study --studyfolder="/public_bme2/bme-zhanghan/Jiayifan/qunex/quickstart"
---> Submitted jobs
NA -> create_study
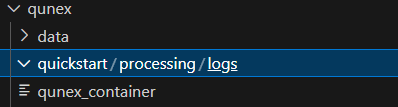
The command does not complete the full build of ‘quickstart’ and the screenshot of the resulting folder is shown below:
So I’m very confused about how to use QuNex correctly on a cluster (hpc), and I’d like to be able to submit the job correctly in order for it to run on the compute node, rather than on the landing node as is suspiciously the case with the third method (which can cause big problems for cluster users).
Your help would be much appreciated! Thank you very much.
Best,
YJia