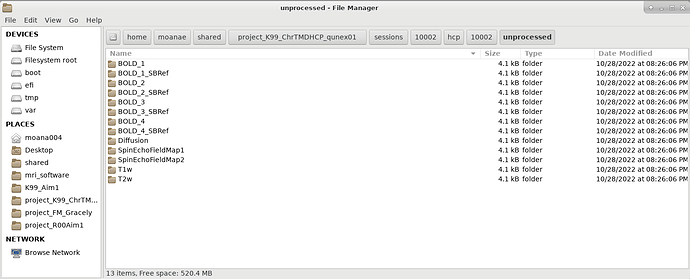
Description:
I’m having troubles when trying to process hcp_fmri_volume, similar to this previous post but in the opposite direction: https://forum.qunex.yale.edu/t/resolved-hcp-fmri-volume-fails-to-detect-automated-naming-scheme-for-bold-image-processing/678. In my case the folders are named BOLD_1, BOLD_2, etc., while qunex looks for BOLD_1_AP, BOLD_2_PA, etc.
A second issue is that using empty quotes for the TOPUP configuration file does not set it to the default, as mentioned in the documentation: https://qunex.readthedocs.io/en/latest/api/gmri/hcp_fmri_volume.html:
–hcp_bold_topupconfig (str, default detailed below):
A full path to the topup configuration file to use. Set to ‘’ if the default is to be used or of TOPUP > distortion correction is not used.
Below is an example runlog file including the errors mentioned above. I also attached the batch file used. Thank you.
Estephan
# Generated by QuNex 0.94.14 on 2022-10-28_16.39.07.866726
#
============================= LOG ================================
# Generated by QuNex 0.94.14 on 2022-10-28_16.39.07.732824
#
=================================================================
gmri hcp_fmri_volume \
--sessionsfolder="/home/moanae/shared/project_K99_ChrTMDHCP_qunex01/sessions" \
--parsessions="52" \
--parelements="4" \
--hcp_filename="automated" \
--sessions="/home/moanae/shared/project_K99_ChrTMDHCP_qunex01/processing/batch_K99Aim2.txt" \
--sessionids="20001,20002,20004,20005,20006,20008,20010,20011,20012,20013,20015,20016,20018,20019,20021,20022,10001,10002,10004,10005,10006,10007,10009,10011,10013,10014,10015,10018,10019,10021,10022,11004,11006,10012,10016,10023,10024,10030,10031,10033,10034,10035,10036,10037,10038,10039,10041,10042,11009,11015" \
=================================================================
Starting multiprocessing sessions in /home/moanae/shared/project_K99_ChrTMDHCP_qunex01/processing/batch_K99Aim2.txt with a pool of 52 concurrent processes
------------------------------------------------------------
Session id: 10016
[started on Friday, 28. October 2022 16:39:07]
Running HCP fMRI Volume pipeline [HCPStyleData] ...
---> PreFS results present.
---> FS results present.
---> PostFS results present.
---> Gradient distortion coefficients file present.
---> Looking for spin echo fieldmap set images [PA/AP].
... identified folder SpinEchoFieldMap1
... phase positive PA spin echo fieldmap image present
... phase negative AP spin echo fieldmap image present
... identified folder SpinEchoFieldMap2
... phase positive PA spin echo fieldmap image present
... phase negative AP spin echo fieldmap image present
---> ERROR: Could not find TOPUP configuration file: ''.
---> Preprocessing settings (unwarpdir, refimage, moveref, seimage) for BOLD 1
... phase encoding direction: AP
... unwarp direction: j-
... using image specific EchoSpacing: 0.000580009 s
... using spin echo fieldmap set 1
-> SE Positive image : 10016_BOLD_PA_SB_SE.nii.gz
-> SE Negative image : 10016_BOLD_AP_SB_SE.nii.gz
... ERROR: bold image missing, searched for ['/home/moanae/shared/project_K99_ChrTMDHCP_qunex01/sessions/10016/hcp/10016/unprocessed/BOLD_1_AP/10016_BOLD_1_AP.nii.gz']!
... ERROR: bold reference image missing!
... using the HCPpipelines default BOLD mask
---> Preprocessing settings (unwarpdir, refimage, moveref, seimage) for BOLD 2
... phase encoding direction: PA
... unwarp direction: j
... using image specific EchoSpacing: 0.000580009 s
... using spin echo fieldmap set 1
-> SE Positive image : 10016_BOLD_PA_SB_SE.nii.gz
-> SE Negative image : 10016_BOLD_AP_SB_SE.nii.gz
... ERROR: bold image missing, searched for ['/home/moanae/shared/project_K99_ChrTMDHCP_qunex01/sessions/10016/hcp/10016/unprocessed/BOLD_2_PA/10016_BOLD_2_PA.nii.gz']!
... ERROR: bold reference image missing!
... using the HCPpipelines default BOLD mask
---> Preprocessing settings (unwarpdir, refimage, moveref, seimage) for BOLD 3
... phase encoding direction: AP
... unwarp direction: j-
... using image specific EchoSpacing: 0.000580009 s
... using spin echo fieldmap set 2
-> SE Positive image : 10016_BOLD_PA_SB_SE.nii.gz
-> SE Negative image : 10016_BOLD_AP_SB_SE.nii.gz
... ERROR: bold image missing, searched for ['/home/moanae/shared/project_K99_ChrTMDHCP_qunex01/sessions/10016/hcp/10016/unprocessed/BOLD_3_AP/10016_BOLD_3_AP.nii.gz']!
... ERROR: bold reference image missing!
... using the HCPpipelines default BOLD mask
---> Preprocessing settings (unwarpdir, refimage, moveref, seimage) for BOLD 4
... phase encoding direction: PA
... unwarp direction: j
... using image specific EchoSpacing: 0.000580009 s
... using spin echo fieldmap set 2
-> SE Positive image : 10016_BOLD_PA_SB_SE.nii.gz
-> SE Negative image : 10016_BOLD_AP_SB_SE.nii.gz
... ERROR: bold image missing, searched for ['/home/moanae/shared/project_K99_ChrTMDHCP_qunex01/sessions/10016/hcp/10016/unprocessed/BOLD_4_PA/10016_BOLD_4_PA.nii.gz']!
... ERROR: bold reference image missing!
... using the HCPpipelines default BOLD mask
Running 4 BOLD images in parallel
---> ERROR: No hcp info for session, skipping this BOLD!
---> ERROR: No hcp info for session, skipping this BOLD!
---> ERROR: No hcp info for session, skipping this BOLD!
---> ERROR: No hcp info for session, skipping this BOLD!
HCP fMRIVolume completed on Friday, 28. October 2022 16:39:07
------------------------------------------------------------
===> Final report for command hcp_fmri_volume
... 10016 ---> HCP fMRI Volume: bolds 1, 2, 3, 4 not ready
===> Not all tasks completed fully!
Call:
subjID_list="20001,20002,20004,20005,20006,20008,20010,20011,20012,20013,20015,20016,20018,20019,20021,20022,10001,10002,10004,10005,10006,10007,10009,10011,10013,10014,10015,10018,10019,10021,10022,11004,11006,10012,10016,10023,10024,10030,10031,10033,10034,10035,10036,10037,10038,10039,10041,10042,11009,11015"; \
msi_resources_time=12:00:00; msi_resources_nodes=1; msi_resources_ntaskspernode=24; msi_resources_mem=64000; msi_queue=agsmall; msi_resources_jobname=K99Aim2_fMRIVolume; \
qunex_container hcp_fmri_volume \
--batchfile=${study_sharedfolder}/processing/batch_K99Aim2.txt --sessionsfolder=${study_sharedfolder}/sessions --sessions=${subjID_list} --parsessions=52 --parelements=4 \
--hcp_filename=automated \
--scheduler=SLURM,array,time=${msi_resources_time},nodes=${msi_resources_nodes},cpus-per-task=${msi_resources_ntaskspernode},mem=${msi_resources_mem},partition=${msi_queue},jobname=${msi_resources_jobname} \
--bind=${study_sharedfolder}:${study_sharedfolder} --container=${HOME}/qunex/qunex_suite-0.94.14.sif
batch_K99Aim2.txt (347.1 KB)