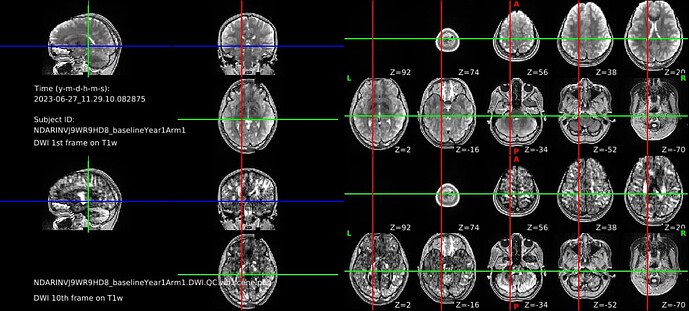
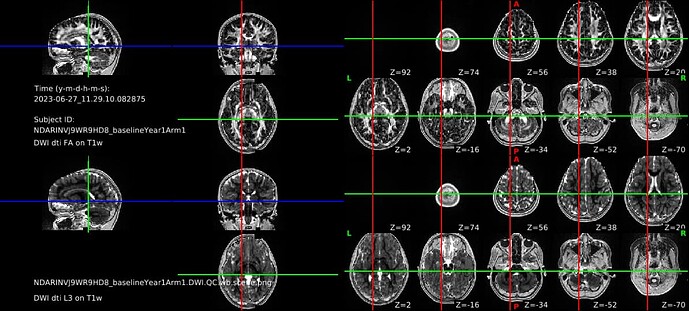
Hello, I’m attempting to run DTIFIT and the tractography pipeline for processed ABCD data downloaded from NDA. The processed data is in bids format, which I renamed and dumped into the Diffusion directory in the hcp/T1w folder (e.g., /gpfs/gibbs/pi/n3/Studies/ABCD/site21/sessions/NDARINVJ9WR9HD8_baselineYear1Arm1/hcp/NDARINVJ9WR9HD8_baselineYear1Arm1/T1w/Diffusion/). I then created the nodif_brain_mask file using outside code (e.g. CBIG/stable_projects/preprocessing/CBIG2022_DiffProc/CBIG_DiffProc_diffusionQC.sh at master · ThomasYeoLab/CBIG · GitHub). I was able to successfully run the qunex DTIFIT command but there is a registration issue (see attached screenshot from the QC command). So, I’m assuming I need to flirt the processed diffusion data (e.g., data.nii.gz) before running the DTIFIT command and I was wondering if there is a QuNex way to do this without having to fully preprocess the data.
Hi Amber,
No unfortunately, there is no such qunex command. I am also not sure what is going here, isn’t ABCD data you are using HCP preprocessed (including diffusion) and should as such be plug and play for DTIFIT? I might be wrong here and this is not the case, in that case I guess you need to run hcp_diffusion on the data first?
Jure