Chiara
September 14, 2022, 2:12pm
1
Hi,parcellate_bold function:
HCP Pipeline :
hcp_pre_freesurfer
hcp_freesurfer
hcp_post_freesurfer
hcp_fmri_volume
hcp_fmri_surface
Mapping and QC
map_hcp_data
run_QC
BOLD task-evoked and resting-state functional connectivity preprocessing
create_bold_brain_masks
compute_bold_stats
create_stats_report
extract_nuisance_signal
preprocess_bold
Once done all of these steps for all of my subjects I’d like to do a connectivity analysis, particularly to assess connectivity between brain parcels. Following what is said here BOLDFunctionalConnectivity , in Resting-state parcellated connectivity paragraph, I’ve tried to launch this command:
qunex parcellate_bold
Now I have a few doubts about some of the parameters to set in this command:
parcellationfile : I can’t find a parcellation file in my /image/functional folder similar to this one proposed in the example (“bold1_Atlas_MSMAll_hp2000_clean.dtseries.nii”), which step should generate it? Which other folder could contain a similar file?
outpath and outname : I’ve tried to set a generic path and name but I riceive an error stating that the parcellation bold file outpath/outname is missing. So these parameters concern an existing file? From the name I thought this setting was to customise the output name and path but for the type of error I’m receveing this “output” should be already somewhere inside my session folder
I’m relatively new to the connectivity analysis so I don’t understand what I’m missing. I’ve already checked on your wiki but it seems to me that there isn’t a step I’ve skipped. Can you help?
Chiara
Hi Chiara,
We are in the process of fixing the documentation for parcellate_bold.
parcellationfile : It should be one of the dlabel files shipped with the qunex container. For example, you can use the Cole Anticevic parcellation using this path (in the container)
/opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii
outpath : Normally the output path is usually <session_id>/images/functional/ which is a folder in each session folder, which means the outpath parameter should be /images/functional/.
outname is a suffix that will be added to the output file name, you can use this parameter to identify the name of the parcellation used to generate the parcellated data.
In addition, the inputfiles parameter should be inputfile. You can find documentation for the command here parcellate_bold — QuNex documentation
Lining
Chiara
September 15, 2022, 1:40pm
3
Hi Lining,
I’ve tried launching the command using your tips but it failed.
This is the full log:
qunex_container parcellate_bold --sessions=/home/admin01/qunex/output/ruminanti_P2/processing/batch_hcp_2use3.txt --sessionsfolder="/home/admin01/qunex/output/ruminanti_P2/sessions" --overwrite=yes --inputfile=bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii --inputpath=/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional --parcellationfile=/opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii --inputdatatype=dtseries --outpath=images/functional --computepconn=yes --outname=parc
(base) admin01@rm-wksfs3:~/qunex$ --> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR PYLIBDIR FSLDIR FSLGPUDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder FSLGPUScripts FSLGPUBinary EDDYCUDADIR USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.93.6
User: root
System: c3d1f586eeb3
OS: RedHat Linux #49~20.04.1-Ubuntu SMP Thu Aug 4 19:15:44 UTC 2022
------------------------------------------------------------------------
██████\ ║ ██\ ██\
██ __██\ ║ ███\ ██ |
██ / ██ |██\ ██\ ║ ████\ ██ | ██████\ ██\ ██\
██ | ██ |██ | ██ | ║ ██ ██\██ |██ __██\\██\ ██ |
██ | ██ |██ | ██ | ║ ██ \████ |████████ |\████ /
██ ██\██ |██ | ██ | ║ ██ |\███ |██ ____|██ ██\
\██████ / \██████ | ║ ██ | \██ |\███████\██ /\██\
\___███\ \______/ ║ \__| \__| \_______\__/ \__|
\___| ║
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSE.md' which is a part of the QuNex Suite source code package:
https://bitbucket.org/oriadev/qunex/src/master/LICENSE.md
---> Setting up Octave
........................ Running QuNex v0.93.6 ........................
Using /home/admin01/qunex/output/ruminanti_P2/processing/batch_hcp_2use3.txt for input.
NOTE: Weights file not used.
Running parcellate_bold with the following parameters:
--------------------------------------------------------------
Study Folder: /home/admin01/qunex/output/ruminanti_P2
Sessions Folder: /home/admin01/qunex/output/ruminanti_P2/sessions
Sessions: 23A
Study Log Folder:
Input File: bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii
Input Path: /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional
Single Input File:
ParcellationFile: /opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii
BOLD Parcellated Connectome Output Name: parc
BOLD Parcellated Connectome Output Path: images/functional
Input Data Type: dtseries
Compute PConn File: yes
Weights file specified to omit certain frames: no
Weights file name: no
Extract data in CSV format: no
Overwrite prior run: yes
--- Full QuNex call for command: parcellate_bold
. /opt/qunex/bash/qx_utilities/parcellate_bold.sh --sessionsfolder='/home/admin01/qunex/output/ruminanti_P2/sessions' --sessions='23A' --inputfile='bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii' --singleinputfile='' --inputpath='/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional' --inputdatatype='dtseries' --parcellationfile='/opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii' --overwrite='yes' --outname='parc' --outpath='images/functional' --computepconn='yes' --extractdata='no' --useweights='no' --weightsfile='no'
--------------------------------------------------------------
--------------------------------------------------------------
Running parcellate_bold locally on c3d1f586eeb3
Command log: /home/admin01/qunex/output/ruminanti_P2/processing/logs/runlogs/Log-parcellate_bold_2022-09-15_09.28.19.839246.log
Command output: /home/admin01/qunex/output/ruminanti_P2/processing/logs/comlogs/tmp_parcellate_bold_23A_2022-09-15_09.28.19.839246.log
--------------------------------------------------------------
-- qunex.sh: Specified Command-Line Options - Start --
SessionsFolder: /home/admin01/qunex/output/ruminanti_P2/sessions
Session: 23A
InputFile: bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii
SingleInputFile:
InputPath: /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional
ParcellationFile: /opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii
OutName: parc
OutPath: images/functional
InputDataType: dtseries
Overwrite: yes
ComputePConn: yes
UseWeights: no
WeightsFile: no
ExtractData: no
-- qunex.sh: Specified Command-Line Options - End --
------------------------- Start of work --------------------------------
--- Establishing paths for all input and output folders:
Working with dtseries files...
Dense BOLD Input: /home/admin01/qunex/output/ruminanti_P2/sessions/23A//home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii
Parcellated BOLD Output: /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii
-- Deleting prior /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii...
-- Checking if parcellation was completed...
-- Note: Prior parcellation data not found because you requested to overwrite.
-- Computing parcellation on /home/admin01/qunex/output/ruminanti_P2/sessions/23A//home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-parcellate /home/admin01/qunex/output/ruminanti_P2/sessions/23A//home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii /opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii COLUMN /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii -only-numeric
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A//home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii', file does not exist, or folder permissions prevent seeing it
-- Using weights: no
-- Computing pconn using correlation...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-correlation /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r.pconn.nii
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii', file does not exist, or folder permissions prevent seeing it
-- Computing GBC using correlation...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-reduce /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r.pconn.nii MEAN /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r_GBC.pscalar.nii -only-numeric
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r.pconn.nii', file does not exist, or folder permissions prevent seeing it
-- Computing pconn using covariance...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-correlation /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_cov.pconn.nii -covariance
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii', file does not exist, or folder permissions prevent seeing it
-- Computing GBC using covariance...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-reduce /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_cov.pconn.nii MEAN /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_cov_GBC.pscalar.nii -only-numeric
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_cov.pconn.nii', file does not exist, or folder permissions prevent seeing it
-- Computing pconn using correlation w/ fisher-z transform...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-correlation /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r_Fz.pconn.nii -fisher-z
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii', file does not exist, or folder permissions prevent seeing it
-- Computing GBC using correlation w/ fisher-z transform...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-reduce /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r_Fz.pconn.nii MEAN /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r_Fz_GBC.pscalar.nii -only-numeric
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r_Fz.pconn.nii', file does not exist, or folder permissions prevent seeing it
-- Checking outputs...
ERROR: Parcellated BOLD file /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii is missing. Something went wrong.
===> ERROR during parcellate_bold. Check final QuNex error log output:
/home/admin01/qunex/output/ruminanti_P2/processing/logs/comlogs/error_parcellate_bold_23A_2022-09-15_09.28.19.839246.log
▄▄▄▄▄▄▄ || ▄▄ ▄▄
▓▓ ▓ || ▓▓▓▄ ▓▓ ▓▓▓▓▓ ▄▓▓▓▓ ▓ ▄▓ ▄▓▓▓ ▓▓▓▄
▓▓ ▓ ▓ ▓ ▓ || ▓▓ ▐▓▓▓ ▄▄▄▄ ▀▓▓ ▓▓▀ ▓▓ ▓ ▓▓ ▓▓ ▓▓ ▄▓▓▄ ▓ ▓▓
▓▓▄▄▓▄▓ ▓▓ ▓ || ▓▓ ▓▓ ▓▄▄▓ ▓▄ ▓▓▀▀ ▓▓▓▓▓ ▓▌ ▓▓ ▀▓▓ ▓ ▓▓
▓▄▄ ▓▓▄▄▓ || ▓▓ ▓ ▓▄▄ ▄▓▓ ▓▓▄ ▓ ▓▀ ▓ ▓ ▓▓▓▀▀ ▓▓▓▓ ▓▓▓▀
||
I also attach the log below:error_parcellate_bold_23A_2022-09-15_09.28.19.839246.log (7.0 KB)
It’s like the function is already searching for an output file inside the folder. Can it be related with some permission error?
Chiara
inputpath should also be images/functional
Chiara
September 15, 2022, 1:54pm
5
Ok, I’ve set inputpath as “image/functional” but I’ve obtained the same error
Can you share your log file again? It will be slightly different. Thanks!
Chiara
September 15, 2022, 1:59pm
7
Sure!
qunex_container parcellate_bold --sessions=/home/admin01/qunex/output/ruminanti_P2/processing/batch_hcp_2use3.txt --sessionsfolder="/home/admin01/qunex/output/ruminanti_P2/sessions" --overwrite=yes --inputfile=bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii --inputpath=images/functional --parcellationfile=/opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii --inputdatatype=dtseries --outpath=images/functional --computepconn=yes --outname=parc
(base) admin01@rm-wksfs3:~/qunex$ --> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR PYLIBDIR FSLDIR FSLGPUDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder FSLGPUScripts FSLGPUBinary EDDYCUDADIR USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.93.6
User: root
System: b9c75b35f743
OS: RedHat Linux #49~20.04.1-Ubuntu SMP Thu Aug 4 19:15:44 UTC 2022
------------------------------------------------------------------------
██████\ ║ ██\ ██\
██ __██\ ║ ███\ ██ |
██ / ██ |██\ ██\ ║ ████\ ██ | ██████\ ██\ ██\
██ | ██ |██ | ██ | ║ ██ ██\██ |██ __██\\██\ ██ |
██ | ██ |██ | ██ | ║ ██ \████ |████████ |\████ /
██ ██\██ |██ | ██ | ║ ██ |\███ |██ ____|██ ██\
\██████ / \██████ | ║ ██ | \██ |\███████\██ /\██\
\___███\ \______/ ║ \__| \__| \_______\__/ \__|
\___| ║
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSE.md' which is a part of the QuNex Suite source code package:
https://bitbucket.org/oriadev/qunex/src/master/LICENSE.md
---> Setting up Octave
........................ Running QuNex v0.93.6 ........................
Using /home/admin01/qunex/output/ruminanti_P2/processing/batch_hcp_2use3.txt for input.
NOTE: Weights file not used.
Running parcellate_bold with the following parameters:
--------------------------------------------------------------
Study Folder: /home/admin01/qunex/output/ruminanti_P2
Sessions Folder: /home/admin01/qunex/output/ruminanti_P2/sessions
Sessions: 23A
Study Log Folder:
Input File: bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii
Input Path: images/functional
Single Input File:
ParcellationFile: /opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii
BOLD Parcellated Connectome Output Name: parc
BOLD Parcellated Connectome Output Path: images/functional
Input Data Type: dtseries
Compute PConn File: yes
Weights file specified to omit certain frames: no
Weights file name: no
Extract data in CSV format: no
Overwrite prior run: yes
--- Full QuNex call for command: parcellate_bold
. /opt/qunex/bash/qx_utilities/parcellate_bold.sh --sessionsfolder='/home/admin01/qunex/output/ruminanti_P2/sessions' --sessions='23A' --inputfile='bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii' --singleinputfile='' --inputpath='images/functional' --inputdatatype='dtseries' --parcellationfile='/opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii' --overwrite='yes' --outname='parc' --outpath='images/functional' --computepconn='yes' --extractdata='no' --useweights='no' --weightsfile='no'
--------------------------------------------------------------
--------------------------------------------------------------
Running parcellate_bold locally on b9c75b35f743
Command log: /home/admin01/qunex/output/ruminanti_P2/processing/logs/runlogs/Log-parcellate_bold_2022-09-15_09.57.09.179474.log
Command output: /home/admin01/qunex/output/ruminanti_P2/processing/logs/comlogs/tmp_parcellate_bold_23A_2022-09-15_09.57.09.179474.log
--------------------------------------------------------------
-- qunex.sh: Specified Command-Line Options - Start --
SessionsFolder: /home/admin01/qunex/output/ruminanti_P2/sessions
Session: 23A
InputFile: bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii
SingleInputFile:
InputPath: images/functional
ParcellationFile: /opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii
OutName: parc
OutPath: images/functional
InputDataType: dtseries
Overwrite: yes
ComputePConn: yes
UseWeights: no
WeightsFile: no
ExtractData: no
-- qunex.sh: Specified Command-Line Options - End --
------------------------- Start of work --------------------------------
--- Establishing paths for all input and output folders:
Working with dtseries files...
Dense BOLD Input: /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii
Parcellated BOLD Output: /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii
-- Deleting prior /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii...
-- Checking if parcellation was completed...
-- Note: Prior parcellation data not found because you requested to overwrite.
-- Computing parcellation on /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-parcellate /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii /opt/qunex/qx_library/data/parcellations/cole_anticevic_net_partition/CortexSubcortex_ColeAnticevic_NetPartition_wSubcorGSR_parcels_LR_ReorderedByNetworks.dlabel.nii COLUMN /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii -only-numeric
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss.dtseries.nii', file does not exist, or folder permissions prevent seeing it
-- Using weights: no
-- Computing pconn using correlation...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-correlation /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r.pconn.nii
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii', file does not exist, or folder permissions prevent seeing it
-- Computing GBC using correlation...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-reduce /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r.pconn.nii MEAN /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r_GBC.pscalar.nii -only-numeric
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r.pconn.nii', file does not exist, or folder permissions prevent seeing it
-- Computing pconn using covariance...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-correlation /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_cov.pconn.nii -covariance
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii', file does not exist, or folder permissions prevent seeing it
-- Computing GBC using covariance...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-reduce /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_cov.pconn.nii MEAN /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_cov_GBC.pscalar.nii -only-numeric
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_cov.pconn.nii', file does not exist, or folder permissions prevent seeing it
-- Computing pconn using correlation w/ fisher-z transform...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-correlation /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r_Fz.pconn.nii -fisher-z
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii', file does not exist, or folder permissions prevent seeing it
-- Computing GBC using correlation w/ fisher-z transform...
While running:
/opt/workbench/workbench-1.5.0/bin_rh_linux64/../exe_rh_linux64/wb_command -cifti-reduce /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r_Fz.pconn.nii MEAN /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r_Fz_GBC.pscalar.nii -only-numeric
ERROR: failed to open file '/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc_r_Fz.pconn.nii', file does not exist, or folder permissions prevent seeing it
-- Checking outputs...
ERROR: Parcellated BOLD file /home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/bold1_Atlas_s_hpss_res-mVWMWBPULSERESP1d_lpss_parc.ptseries.nii is missing. Something went wrong.
===> ERROR during parcellate_bold. Check final QuNex error log output:
/home/admin01/qunex/output/ruminanti_P2/processing/logs/comlogs/error_parcellate_bold_23A_2022-09-15_09.57.09.179474.log
▄▄▄▄▄▄▄ || ▄▄ ▄▄
▓▓ ▓ || ▓▓▓▄ ▓▓ ▓▓▓▓▓ ▄▓▓▓▓ ▓ ▄▓ ▄▓▓▓ ▓▓▓▄
▓▓ ▓ ▓ ▓ ▓ || ▓▓ ▐▓▓▓ ▄▄▄▄ ▀▓▓ ▓▓▀ ▓▓ ▓ ▓▓ ▓▓ ▓▓ ▄▓▓▄ ▓ ▓▓
▓▓▄▄▓▄▓ ▓▓ ▓ || ▓▓ ▓▓ ▓▄▄▓ ▓▄ ▓▓▀▀ ▓▓▓▓▓ ▓▌ ▓▓ ▀▓▓ ▓ ▓▓
▓▄▄ ▓▓▄▄▓ || ▓▓ ▓ ▓▄▄ ▄▓▓ ▓▓▄ ▓ ▓▀ ▓ ▓ ▓▓▓▀▀ ▓▓▓▓ ▓▓▓▀
||
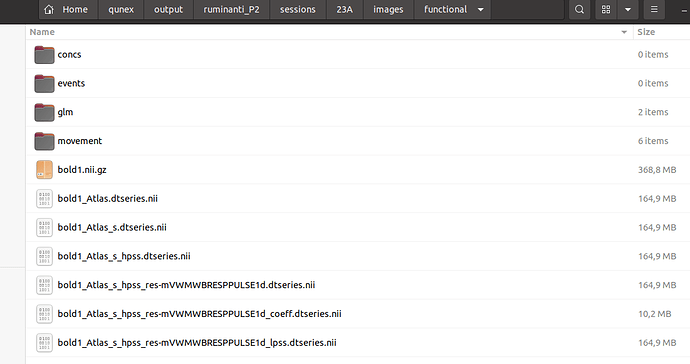
Can you show me the content of this folder?/home/admin01/qunex/output/ruminanti_P2/sessions/23A/images/functional/
The actual file name depends on the parameters used during preprocess bold.
Chiara
September 15, 2022, 2:12pm
9
This is the content of /image/functional:
Can you double check your input file name? It should be bold1_Atlas_s_hpss_res-mVWMWBRESPPULSE1d_lpss.dtseries.nii in your case.
1 Like
Chiara
September 15, 2022, 2:47pm
11
Ok, now it has gone correctly. There was a typo in the input file name.
No problem. That’s great!
1 Like
Hi @Chiara I’m wondering if you were ultimately able to run palm stats on your parcellated rFC data?
Chiara
October 12, 2022, 1:07pm
14
Hi @rachael_grazioplene ,
Sorry for the late response. Unfortunately, I have not yet done any palm analysis.
Chiara
Yeah, I’m not quite sure what the trouble is. I’ll make a separate post. Thanks for your reply!