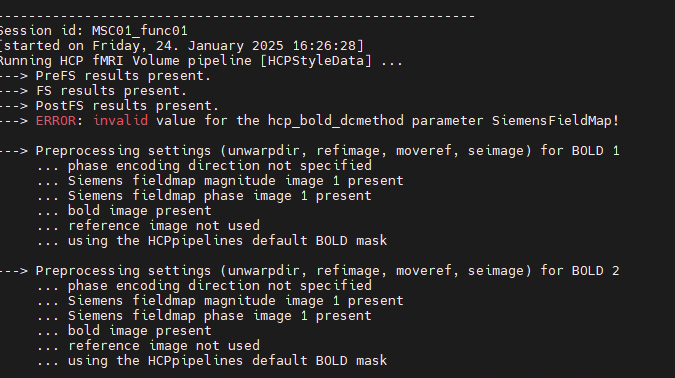
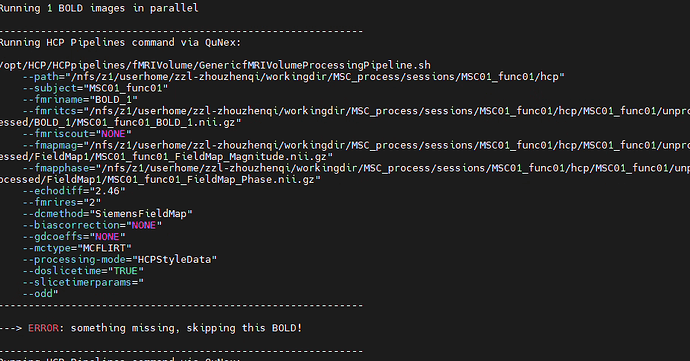
Hi, I’m a new user of Qunex. I’m trying to process the MSC dataset using HCPpipeline. Each subject has multiple sessions, and the data is in BIDS format. However, the functional and structural sessions are separated. Each subject has two structural sessions, while the remaining functional sessions only contain func and fmap data. How should I handle this situation?
The following is a portion of the folder structure for one of the subjects.
├── ses-func09
│ ├── fmap
│ │ ├── sub-MSC01_ses-func09_magnitude1.nii.gz
│ │ ├── sub-MSC01_ses-func09_magnitude2.nii.gz
│ │ └── sub-MSC01_ses-func09_phasediff.nii.gz
│ ├── func
│ │ ├── sub-MSC01_ses-func09_task-glasslexical_run-01_bold.nii.gz
│ │ ├── sub-MSC01_ses-func09_task-glasslexical_run-01_events.tsv
│ │ ├── sub-MSC01_ses-func09_task-glasslexical_run-02_bold.nii.gz
│ │ ├── sub-MSC01_ses-func09_task-glasslexical_run-02_events.tsv
│ │ ├── sub-MSC01_ses-func09_task-memoryfaces_bold.nii.gz
│ │ ├── sub-MSC01_ses-func09_task-memoryfaces_events.tsv
│ │ ├── sub-MSC01_ses-func09_task-memoryscenes_bold.nii.gz
│ │ ├── sub-MSC01_ses-func09_task-memoryscenes_events.tsv
│ │ ├── sub-MSC01_ses-func09_task-memorywords_bold.nii.gz
│ │ ├── sub-MSC01_ses-func09_task-memorywords_events.tsv
│ │ ├── sub-MSC01_ses-func09_task-motor_run-01_bold.nii.gz
│ │ ├── sub-MSC01_ses-func09_task-motor_run-01_events.tsv
│ │ ├── sub-MSC01_ses-func09_task-motor_run-02_bold.nii.gz
│ │ ├── sub-MSC01_ses-func09_task-motor_run-02_events.tsv
│ │ └── sub-MSC01_ses-func09_task-rest_bold.nii.gz
│ └── sub-MSC01_ses-func09_scans.tsv
├── ses-func10
│ ├── fmap
│ │ ├── sub-MSC01_ses-func10_magnitude1.nii.gz
│ │ ├── sub-MSC01_ses-func10_magnitude2.nii.gz
│ │ └── sub-MSC01_ses-func10_phasediff.nii.gz
│ ├── func
│ │ ├── sub-MSC01_ses-func10_task-glasslexical_run-01_bold.nii.gz
│ │ ├── sub-MSC01_ses-func10_task-glasslexical_run-01_events.tsv
│ │ ├── sub-MSC01_ses-func10_task-glasslexical_run-02_bold.nii.gz
│ │ ├── sub-MSC01_ses-func10_task-glasslexical_run-02_events.tsv
│ │ ├── sub-MSC01_ses-func10_task-memoryfaces_bold.nii.gz
│ │ ├── sub-MSC01_ses-func10_task-memoryfaces_events.tsv
│ │ ├── sub-MSC01_ses-func10_task-memoryscenes_bold.nii.gz
│ │ ├── sub-MSC01_ses-func10_task-memoryscenes_events.tsv
│ │ ├── sub-MSC01_ses-func10_task-memorywords_bold.nii.gz
│ │ ├── sub-MSC01_ses-func10_task-memorywords_events.tsv
│ │ ├── sub-MSC01_ses-func10_task-motor_run-01_bold.nii.gz
│ │ ├── sub-MSC01_ses-func10_task-motor_run-01_events.tsv
│ │ ├── sub-MSC01_ses-func10_task-motor_run-02_bold.nii.gz
│ │ ├── sub-MSC01_ses-func10_task-motor_run-02_events.tsv
│ │ └── sub-MSC01_ses-func10_task-rest_bold.nii.gz
│ └── sub-MSC01_ses-func10_scans.tsv
├── ses-struct01
│ ├── anat
│ │ ├── sub-MSC01_ses-struct01_run-01_angio.nii.gz
│ │ ├── sub-MSC01_ses-struct01_run-01_mod-angio_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct01_run-01_mod-T1w_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct01_run-01_mod-T2w_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct01_run-01_T1w.nii.gz
│ │ ├── sub-MSC01_ses-struct01_run-01_T2w.nii.gz
│ │ ├── sub-MSC01_ses-struct01_run-02_angio.nii.gz
│ │ ├── sub-MSC01_ses-struct01_run-02_mod-angio_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct01_run-02_mod-T1w_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct01_run-02_mod-T2w_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct01_run-02_T1w.nii.gz
│ │ └── sub-MSC01_ses-struct01_run-02_T2w.nii.gz
│ └── sub-MSC01_ses-struct01_scans.tsv
├── ses-struct02
│ ├── anat
│ │ ├── sub-MSC01_ses-struct02_acq-coronal_run-01_mod-veno_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-coronal_run-01_veno.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-coronal_run-02_mod-veno_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-coronal_run-02_veno.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-coronal_run-03_mod-veno_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-coronal_run-03_veno.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-sagittal_run-01_mod-veno_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-sagittal_run-01_veno.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-sagittal_run-02_mod-veno_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-sagittal_run-02_veno.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-sagittal_run-03_mod-veno_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-sagittal_run-03_veno.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-sagittal_run-04_mod-veno_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_acq-sagittal_run-04_veno.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-01_angio.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-01_mod-angio_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-01_mod-T1w_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-01_mod-T2w_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-01_T1w.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-01_T2w.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-02_angio.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-02_mod-angio_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-02_mod-T1w_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-02_mod-T2w_defacemask.nii.gz
│ │ ├── sub-MSC01_ses-struct02_run-02_T1w.nii.gz
│ │ └── sub-MSC01_ses-struct02_run-02_T2w.nii.gz
│ └── sub-MSC01_ses-struct02_scans.tsv
Do I need to manually adjust the paths of the structural image folders before running import_bids? Or should I configure certain settings in one of the steps after import_bids?