Hi there,
I am trying to process multimodal MRI data using HCPpipeline under Qunex(qunex_suite-0.93.2.sif). My bids data and process scrpts are as follows:
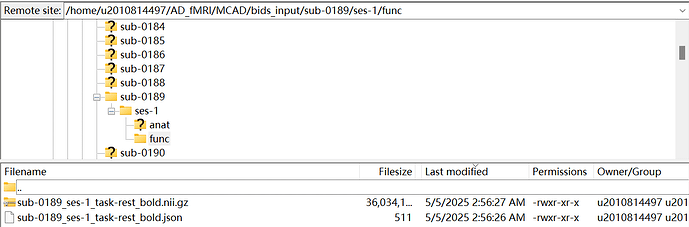
Bids data:
Batch script(rename sh file to txt for upload):
Step2_QuNexSingularity_sbatch.txt (1.6 KB)
Qunex script(rename sh file to txt for upload):
QuNexSingularity_Commands_sll.txt (3.8 KB)
Batch file:
batch_MCAD.txt (3.6 KB)
Mapping file:
mapping_MCAD.txt (97 Bytes)
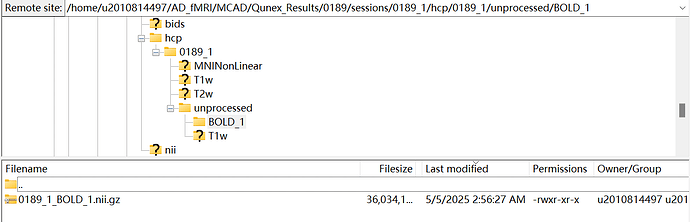
Now the problem is that this pipeline works well with T1 data, but can not recongize fMRI data. I’ve checked the output in /home/u2010814497/AD_fMRI/MCAD/Qunex_Results/0189/sessions/0189_1/hcp/0189_1, there is no BOLD related subfolder, I can only find them in /home/u2010814497/AD_fMRI/MCAD/Qunex_Results/0189/sessions/0189_1/hcp/0189_1/unprocessed as shown in the following picture:
The output log in /home/u2010814497/AD_fMRI/MCAD/Qunex_Results/0189/processing/logs/comlogs showed done for create_study, import_bids, create_session_info, setup_hcp, create_batch, hcp_pre_freesurfer, hcp_freesurfer, hcp_post_freesurfer. But can not run hcp_fmri_volume and hcp_fmri_surface due to missing data in /home/u2010814497/AD_fMRI/MCAD/Qunex_Results/0189/sessions/0189_1/hcp/0189_1/MNINonLinear/Results/BOLD_1.
The batch file generated for subject 0189 were as follows:
batch.txt (4.5 KB)
It seems that something is wrong with create_session_info for fmri data. As a beginner, I don’t know how to debug this problem. I was wondering if I can get help for this.
Looking forward to your reply, best wishes!