I am trying to do dry run of the HCP PreFreeSurfer but I am running into an issue of qunex not finding the batch file I created. I tried using absolute and relative paths, and they lead to two different errors: 1. cannot find the batch file; 2. cannot find a log file, respectively. See the terminal output below. Please advise, thank you.
cn2107:~ moana004$ study_sharedfolder="/home/moanae/shared/project_FM_Gracely_derivatives"
cn2107:~ moana004$ ls ${study_sharedfolder}/processing/batch_01_structural.txt
/home/moanae/shared/project_FM_Gracely_derivatives/processing/batch_01_structural.txt
cn2107:~ moana004$ qunex_container hcp_pre_freesurfer --batchfile="/FM_Gracely_shared/processing/batch_01_structural.txt" --sessionsfolder="/FM_Gracely_shared/sessions" --test --bind="${study_sharedfolder}:/FM_Gracely_shared" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
Traceback (most recent call last):
File "/home/moanae/moana004/qunex/qunex_container", line 1118, in <module>
main()
File "/home/moanae/moana004/qunex/qunex_container", line 904, in main
g_sessionids = get_sessionids(batchfile, sessions, batchfilter)
File "/home/moanae/moana004/qunex/qunex_container", line 467, in get_sessionids
raise ValueError("ERROR: The specified file is not found! [%s]!" % batchfile)
ValueError: ERROR: The specified file is not found! [/FM_Gracely_shared/processing/batch_01_structural.txt]!
cn2107:~ moana004$ cd ${study_sharedfolder}; qunex_container hcp_pre_freesurfer --batchfile="processing/batch_01_structural.txt" --sessionsfolder="sessions" --test --bind="${study_sharedfolder}:/FM_Gracely_shared" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
--> QuNex will run the command over 2 sessions. It will utilize:
Maximum sessions run in parallel for a job: 1.
Maximum elements run in parallel for a session: 1.
Up to 1 processes will be utilized for a job.
Job #1 will run sessions: ,FM0001
cn2107:project_FM_Gracely_derivatives moana004$ /usr/bin/bash: /panfs/roc/groups/12/moanae/shared/project_FM_Gracely_derivatives/processing/logs/batchlogs/qunex_container_command_2022-08-08_13.55.58.717858.sh: No such file or directory
Hi, Estephan!
The problem here is that the qunex_container script operates outside of the container to setup parallelism over sessions and as such needs access to the batch file as well. Now it is looking for the batch file in the path that is valid once inside the container. This is a tricky thing to patch and we might need to think this through a bit more. The easiest solution for you for the time being is to just bind path inside the container to the same ones that are used outside, so:
qunex_container hcp_pre_freesurfer \
--batchfile="${study_sharedfolder}/processing/batch_01_structural.txt" \
--sessionsfolder="${study_sharedfolder}/sessions" \
--bind="${study_sharedfolder}:/${study_sharedfolder}" \
--container="${HOME}/qunex/qunex_suite-0.94.4.sif"
Let me know if this works, thanks!
This worked to get qunex_container to process the command for a single session. However the dry run complains of not finding the T1w image file.
I attached below the command output, the “bis2nii.log” and the batch file I tried. Please note that I had to fiddle a bit the steps to prepare the batch file for this study, as my MRI legacy data has a different imaging sequence than the HCP protocol (and it is my first attempt in using QuNex). Can you help me find the issue? Thank you.
Command output
cn2107:project_FM_Gracely_derivatives moana004$ qunex_container hcp_pre_freesurfer --batchfile="${study_sharedfolder}/processing/batch_01_structural.txt" --sessionsfolder="${study_sharedfolder}/sessions" --test --bind="${study_sharedfolder}:/${study_sharedfolder}" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
--> QuNex will run the command over 2 sessions. It will utilize:
Maximum sessions run in parallel for a job: 1.
Maximum elements run in parallel for a session: 1.
Up to 1 processes will be utilized for a job.
Job #1 will run sessions: ,FM0001
cn2107:project_FM_Gracely_derivatives moana004$ --> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR ANTSDIR PYLIBDIR FSLDIR FSLGPUDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder FSL_GPU_SCRIPTS FSLGPUBinary EDDYCUDADIR USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.94.4
User: moana004
System: cn2107
OS: RedHat Linux #1 SMP Tue Jun 28 15:37:28 UTC 2022
------------------------------------------------------------------------
ââââââ\ â ââ\ ââ\
ââ __ââ\ â âââ\ ââ |
ââ / ââ |ââ\ ââ\ â ââââ\ ââ | ââââââ\ ââ\ ââ\
ââ | ââ |ââ | ââ | â ââ ââ\ââ |ââ __ââ\\ââ\ ââ |
ââ | ââ |ââ | ââ | â ââ \ââââ |ââââââââ |\ââââ /
ââ ââ\ââ |ââ | ââ | â ââ |\âââ |ââ ____|ââ ââ\
\ââââââ / \ââââââ | â ââ | \ââ |\âââââââ\ââ /\ââ\
\___âââ\ \______/ â \__| \__| \_______\__/ \__|
\___| â
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSES' which is a part of the QuNex Suite source code package:
https://gitlab.qunex.yale.edu/qunex/qunex/-/tree/master/LICENSES
---> Setting up Octave
........................ Running QuNex v0.94.4 ........................
--- Full QuNex call for command: hcp_pre_freesurfer
gmri hcp_pre_freesurfer --sessionsfolder="/home/moanae/shared/project_FM_Gracely_derivatives/sessions" --test --batchfile="/home/moanae/shared/project_FM_Gracely_derivatives/processing/batch_01_structural.txt" --sessions=",FM0001"
---------------------------------------------------------
WARNING: Parameter qx_cifti_tail was not specified. Its value was imputed from parameter hcp_cifti_tail and set to ''!
WARNING: Parameter qx_nifti_tail was not specified. Its value was imputed from parameter hcp_nifti_tail and set to ''!
WARNING: Parameter cifti_tail was not specified. Its value was imputed from parameter qx_cifti_tail and set to ''!
WARNING: Parameter nifti_tail was not specified. Its value was imputed from parameter qx_nifti_tail and set to ''!
# Generated by QuNex 0.94.4 on 2022-08-08_17.26.02.654445
#
=================================================================
gmri hcp_pre_freesurfer \
--sessionsfolder="/home/moanae/shared/project_FM_Gracely_derivatives/sessions" \
--test="True" \
--sessions="/home/moanae/shared/project_FM_Gracely_derivatives/processing/batch_01_structural.txt" \
--sessionids=",FM0001" \
=================================================================
Running test on /home/moanae/shared/project_FM_Gracely_derivatives/processing/batch_01_structural.txt ...
---- Running local
Starting testing of sessions FM0001 at Monday, 08. August 2022 17:26:02
------------------------------------------------------------
Session id: FM0001
[started on Monday, 08. August 2022 17:26:02]
Test running HCP PreFreeSurfer Pipeline [LegacyStyleData] ...
---> ERROR: Could not find T1w image file. []
---> Not using T2w image.
---> WARNING: No distortion correction method specified.
---> Due to missing files session can not be processed.
HCP PreFS test completed on Monday, 08. August 2022 17:26:02
------------------------------------------------------------
===> Final report for command hcp_pre_freesurfer
... FM0001 ---> Files missing, PreFS can not be run
===> Not all tasks completed fully!
bis2nii.log
BIDS to nii mapping report, executed on 2022-08-06T22:28:30
/FM_Gracely_shared/sessions/FM0001/bids/anat/sub-FM0001_run-01_PDw.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/1.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/anat/sub-FM0001_run-01_T1w.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/2.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/anat/sub-FM0001_run-01_T2w.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/3.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-asl_run-01_phasediff.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/4.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-asl_run-01_magnitude2.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/5.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-asl_run-01_magnitude1.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/6.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-dwi_run-01_phasediff.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/7.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-dwi_run-01_magnitude2.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/8.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-dwi_run-01_magnitude1.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/9.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-funcrest_run-01_magnitude1.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/10.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-funcrest_run-01_magnitude2.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/11.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-funcrest_run-01_phasediff.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/12.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-funcstim_run-01_magnitude1.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/13.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-funcstim_run-01_magnitude2.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/14.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/fmap/sub-FM0001_acq-funcstim_run-01_phasediff.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/15.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/func/sub-FM0001_task-auditory_run-03_bold.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/16.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/func/sub-FM0001_task-intermingled_run-01_bold.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/17.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/func/sub-FM0001_task-mechanical_run-02_bold.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/18.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/func/sub-FM0001_task-rest_run-01_bold.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/19.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/func/sub-FM0001_task-rest_run-02_bold.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/20.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/func/sub-FM0001_task-simultaneous_run-04_bold.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/21.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/dwi/sub-FM0001_run-01_dwi.nii.gz => /FM_Gracely_shared/sessions/FM0001/nii/22.nii.gz
/FM_Gracely_shared/sessions/FM0001/bids/dwi/sub-FM0001_run-01_dwi.bvec => /FM_Gracely_shared/sessions/FM0001/nii/22.bvec
/FM_Gracely_shared/sessions/FM0001/bids/dwi/sub-FM0001_run-01_dwi.bval => /FM_Gracely_shared/sessions/FM0001/nii/22.bval
Batch file
batch_01_structural.txt (41.7 KB)
A side note: during the “import_bids” step, it seems that the ASL data was ignored and it was no imported to the “nii” folder. Is this an expected behavior?
Sure.
### Preparing the qunex folder structure
## Starting up a study involves only creating the basic folder structure that will be used by QuNex commands and functions
# Variables needed
study_sharedfolder="/home/moanae/shared/project_FM_Gracely_derivatives"
# Create folder in shared (needed once, before binding it for singularity)
mkdir -p ${study_sharedfolder}
# qunex command (need to bind any folders not included by default by singularity, such as $HOME)
qunex_container create_study --bind="${study_sharedfolder}:/FM_Gracely_shared" --studyfolder="/FM_Gracely_shared" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
###
### Onboard BIDS-formatted data to qunex
# Variables needed
study_sharedfolder="/home/moanae/shared/project_FM_Gracely_derivatives"; BIDSorig="${HOME}/project_FM_Gracely/BIDS_output"; BIDSqunex=${study_sharedfolder}/sessions/inbox/BIDS
# Manual copy of BIDS_output folder to qunex
cp -R ${BIDSorig}/* ${BIDSqunex}
# First attempt with one participant data (sub-FM0001)
# qunex command (need to bind any folders not included by default by singularity, such as $HOME)
qunex_container import_bids --bind="${study_sharedfolder}:/FM_Gracely_shared" --sessionsfolder="/FM_Gracely_shared/sessions" --sessions='sub-FM0001' --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
# All other participants
# Variable with list of participants to be processed, removes sub-FM0001 (bash command tr used to remove newlines)
subjs_list=`ls -d ${BIDSorig}/sub-?????? | awk -F/ '{print $NF ","}' | sed 's/sub-FM0001,//' | tr '\n' ' '`; qunex_container import_bids --bind="${study_sharedfolder}:/FM_Gracely_shared" --sessionsfolder="/FM_Gracely_shared/sessions" --sessions="${subjs_list}" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
## Had to manually remove the folder "HC0436func", since it is an empty folder but still recorded in the BIDS conversion done as described in file FM_Gracely_DataAnalysisCommands_01_dicom2nifti.txt
###
# Command for qunex_container create_session_info: v0.93.6
qunex_container create_session_info --pipelines=hcp --sessionsfolder="/FM_Gracely_shared/sessions" --sourcefile=session.txt --targetfile=session_hcp.txt --mapping=/FM_Gracely_shared/sessions/specs/hcp_mapping.txt --bind="${study_sharedfolder}:/FM_Gracely_shared" --container="${HOME}/qunex/qunex_suite-0.93.6.sif"
## NOTE: qunex is limited in how it handles multiple field map files. There is a need for manual manipulation of the session_mod_hcp.txt files to have the field maps numbered in the order that they are supposed to be used. For example, the field maps for task-bold should have the same fm(x) number as the task-bold runs.
## Created a manually modified session_hcp2.txt file for each participant by looking at the imaging sequence described in the PACS data file.
# Decided to use the following convention: funcrest: fm(1), funcstim: fm(2); dwi: fm(3)
# If more than one for each type, add a second digit: fm(11), fm(12)
## ASL filed maps not processed, as qunex cannot handle ASL data files apparently.
###
### Mapping the files into the HCP file structure
item="FM0001"; qunex_container setup_hcp --sourcefolder="/FM_Gracely_shared/sessions/${item}" --targetfolder='hcp' --sourcefile=session_hcp2.txt --check='yes' --existing='add' --hcp_filename='userdefined' --hcp_folderstructure='hcpls' --bind="${study_sharedfolder}:/FM_Gracely_shared" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
###
## prepare a batch file using minimum parameters for running HCP pipelines
item="FM0001"; qunex_container create_batch --sessions="${item}" --sessionsfolder="/FM_Gracely_shared/sessions" --sourcefiles=session_hcp2.txt --targetfile="/FM_Gracely_shared/processing/batch_01_structural.txt" --paramfile="/FM_Gracely_shared/sessions/specs/parameters_01_structural.txt" --bind="${study_sharedfolder}:/FM_Gracely_shared" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
Mapping file
hcp_mapping.txt (4.0 KB)
- Is this what you would like to see?
cn2107:~ moana004$ ls /home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/*
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0001:
anat dwi fmap func perf sub-FM0001_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0007:
anat dwi fmap func perf sub-FM0007_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0097:
anat dwi fmap func perf sub-FM0097_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0530:
anat dwi fmap func perf sub-FM0530_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0608:
anat dwi fmap func perf sub-FM0608_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0663:
anat dwi fmap func perf sub-FM0663_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0757:
anat dwi fmap func perf sub-FM0757_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0761:
anat dwi fmap func perf sub-FM0761_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0765:
anat dwi fmap func perf sub-FM0765_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0770:
anat dwi fmap func perf sub-FM0770_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0785:
anat dwi fmap func perf sub-FM0785_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0796:
anat dwi fmap func perf sub-FM0796_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0797:
anat dwi fmap func perf sub-FM0797_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0804:
anat dwi fmap func perf sub-FM0804_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0826:
anat dwi fmap func perf sub-FM0826_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0833:
anat dwi fmap func perf sub-FM0833_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0867:
anat dwi fmap func perf sub-FM0867_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM1071:
anat dwi fmap func perf sub-FM1071_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2001:
anat dwi fmap func perf sub-FM2001_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2162:
anat dwi fmap func perf sub-FM2162_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2382:
anat dwi fmap func perf sub-FM2382_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2383:
anat dwi fmap func perf sub-FM2383_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2388:
anat dwi fmap func perf sub-FM2388_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2439:
anat dwi fmap func perf sub-FM2439_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2443:
anat dwi fmap func perf sub-FM2443_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2451:
anat dwi fmap func perf sub-FM2451_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2465:
anat dwi fmap func perf sub-FM2465_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2473:
anat dwi fmap func perf sub-FM2473_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2474:
anat dwi fmap func perf sub-FM2474_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM2526:
anat dwi fmap func perf sub-FM2526_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0002:
anat dwi fmap func perf sub-HC0002_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0003:
anat dwi fmap func perf sub-HC0003_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0068:
anat dwi fmap func perf sub-HC0068_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0102:
anat dwi fmap func perf sub-HC0102_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0182:
anat dwi fmap func perf sub-HC0182_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0229:
anat dwi fmap func perf sub-HC0229_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0249:
anat dwi fmap func perf sub-HC0249_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0267:
anat dwi fmap func perf sub-HC0267_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0287:
anat dwi fmap func perf sub-HC0287_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0293:
anat dwi fmap func perf sub-HC0293_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0303:
anat dwi fmap func perf sub-HC0303_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0308:
anat dwi fmap func perf sub-HC0308_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0436:
anat dwi fmap func perf sub-HC0436func_scans.tsv sub-HC0436_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0560:
anat dwi fmap func perf sub-HC0560_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0588:
anat dwi fmap func perf sub-HC0588_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0610:
anat dwi fmap func perf sub-HC0610_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0782:
anat dwi fmap func perf sub-HC0782_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC0813:
anat dwi fmap func perf sub-HC0813_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2054:
anat dwi fmap func perf sub-HC2054_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2062:
anat dwi fmap func perf sub-HC2062_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2105:
anat dwi fmap func perf sub-HC2105_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2106:
anat dwi fmap func perf sub-HC2106_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2128:
anat dwi fmap func perf sub-HC2128_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2246:
anat dwi fmap func perf sub-HC2246_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2277:
anat dwi fmap func perf sub-HC2277_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2284:
anat dwi fmap func perf sub-HC2284_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2422:
anat dwi fmap func perf sub-HC2422_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2436:
anat dwi fmap func perf sub-HC2436_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2501:
anat dwi fmap func perf sub-HC2501_scans.tsv
/home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-HC2515:
anat dwi fmap func perf sub-HC2515_scans.tsv
Hi Estephan,
Thanks for providing the commands and the folder structure!
What we really want to see is the content of the folder that contains asl scans.
- If you happen to have the
tree command available, could you run tree command on one of the subjects.
tree /home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS/sub-FM0001
- If not, showing us the content of the
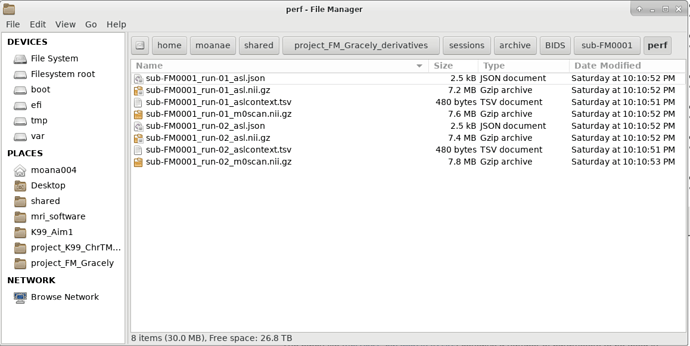
perf folder should be enough.
Lining
Got it. I don’t seem to have the ‘tree’ command available in my HPC cluster, so here is a screen shoot of the ‘perf’ folder.
Hi Estephan,
I believe the issue is that you are not using consistent bindings throughout commands. As a result the paths in the batch file are messed up.
For example, when running hcp_pre_freesurfer, you are providing the paths as /home/moanae/shared/project_FM_Gracely_derivatives/sessions while in the batch file the paths are stored as /FM_Gracely_shared/sessions which confuses QuNex. Just rerun all the initial commands by using the same binding as it will be later in hcp_pre_freesurfer and you should be good to go. Just always use the binding:
--bind="${study_sharedfolder}:/${study_sharedfolder}"
Here it is what I tried, starting from scratch:
# Variables needed
study_sharedfolder="/home/moanae/shared/project_FM_Gracely_derivatives"; BIDSorig="${HOME}/project_FM_Gracely/BIDS_output"; BIDSqunex=${study_sharedfolder}/sessions/inbox/BIDS
# Create folder in shared (needed once, before binding it for singularity)
mkdir -p ${study_sharedfolder}
# qunex command (need to bind any folders not included by default by singularity, such as $HOME)
qunex_container create_study --studyfolder="${study_sharedfolder}" --bind="${study_sharedfolder}:/${study_sharedfolder}" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
# Manual copy of BIDS_output folder to qunex
cp -R ${BIDSorig}/* ${BIDSqunex}
# qunex command (need to bind any folders not included by default by singularity, such as $HOME)
qunex_container import_bids --sessionsfolder="${study_sharedfolder}/sessions" --sessions='sub-FM0001' --bind="${study_sharedfolder}:/${study_sharedfolder}" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
The last command spits out this error:
cn2107:~ moana004$ qunex_container import_bids --sessionsfolder="${study_sharedfolder}/sessions" --sessions='sub-FM0001' --bind="${study_sharedfolder}:/${study_sharedfolder}" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
--> QuNex will run the command over 1 sessions. It will utilize:
Maximum sessions run in parallel for a job: 1.
Maximum elements run in parallel for a session: 1.
Up to 1 processes will be utilized for a job.
Job #1 will run sessions: sub-FM0001
cn2107:~ moana004$ --> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR ANTSDIR PYLIBDIR FSLDIR FSLGPUDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder FSL_GPU_SCRIPTS FSLGPUBinary EDDYCUDADIR USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.94.4
User: moana004
System: cn2107
OS: RedHat Linux #1 SMP Tue Jun 28 15:37:28 UTC 2022
------------------------------------------------------------------------
ââââââ\ â ââ\ ââ\
ââ __ââ\ â âââ\ ââ |
ââ / ââ |ââ\ ââ\ â ââââ\ ââ | ââââââ\ ââ\ ââ\
ââ | ââ |ââ | ââ | â ââ ââ\ââ |ââ __ââ\\ââ\ ââ |
ââ | ââ |ââ | ââ | â ââ \ââââ |ââââââââ |\ââââ /
ââ ââ\ââ |ââ | ââ | â ââ |\âââ |ââ ____|ââ ââ\
\ââââââ / \ââââââ | â ââ | \ââ |\âââââââ\ââ /\ââ\
\___âââ\ \______/ â \__| \__| \_______\__/ \__|
\___| â
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSES' which is a part of the QuNex Suite source code package:
https://gitlab.qunex.yale.edu/qunex/qunex/-/tree/master/LICENSES
---> Setting up Octave
........................ Running QuNex v0.94.4 ........................
--- Full QuNex call for command: import_bids
gmri import_bids --sessionsfolder="/home/moanae/shared/project_FM_Gracely_derivatives/sessions" --sessions="sub-FM0001"
---------------------------------------------------------
started running import_bids at 2022-08-09 11:47:25, track progress in /home/moanae/shared/project_FM_Gracely_derivatives/processing/logs/comlogs/tmp_import_bids_sub-FM0001_2022-08-09_11.47.25.332431.log
call: gmri import_bids sessionsfolder="/home/moanae/shared/project_FM_Gracely_derivatives/sessions" sessions="sub-FM0001"
-----------------------------------------
Running import_bids
==================
--> identifying files in /home/moanae/shared/project_FM_Gracely_derivatives/sessions/inbox/BIDS
--> Inbox type: bids_study
==> Paths:
BIDSInfo -> /home/moanae/shared/project_FM_Gracely_derivatives/info/bids
BIDSInbox -> /home/moanae/shared/project_FM_Gracely_derivatives/sessions/inbox/BIDS
BIDSArchive -> /home/moanae/shared/project_FM_Gracely_derivatives/sessions/archive/BIDS
--> mapping files to QuNex bids folders
--> processing BIDS info folder
ERROR
Traceback (most recent call last):
File "/opt/qunex/python/qx_utilities/general/core.py", line 570, in runWithLog
result = function(**args)
File "/opt/qunex/python/qx_utilities/general/bids.py", line 600, in import_bids
tfile, lock = mapToQUNEXBids(file, sessionsfolder, BIDSInfo, sessionsList, overwrite, " ")
File "/opt/qunex/python/qx_utilities/general/bids.py", line 160, in mapToQUNEXBids
fparts = [folder] + oparts[oparts.index(optional):]
ValueError: 'derivatives' is not in list
-----------------------------------------
Finished at 2022-08-09 11:47:25
It seems that the idea of using a subfolder in the “shared” folder seems quite a hassle. My intnetion was to allow other members of my group to access these files (if the live in my $HOME folder, they would not have access to it). Is this worth pursuing or should I just run everything in my $HOME folder? Thank you.
There shouldn’t be any issues regarding shared files, things should work normally. The problem right now is that you are using a keyword derivatives in your study name, derivatives is a BIDS keyword and as such confuses the BIDS importer. Can you please try picking a different study name, say project_FM.
In the next version, we will make QuNex more robust by looking for BIDS compliance only within the folders inside the study and not in the full path.
Thanks for the feedback, your suggestion helped me move forward and I was able to call the hcp_pre_freesurfer command successfully. However it did stop early due to an error regarding file permissions, see log file below. How should I proceed?
qunex_container command
cn2107:~ moana004$ qunex_container hcp_pre_freesurfer --batchfile="${study_sharedfolder}/processing/batch_01_structural.txt" --sessionsfolder="${study_sharedfolder}/sessions" --bind="${study_sharedfolder}:/${study_sharedfolder}" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
--> QuNex will run the command over 2 sessions. It will utilize:
Maximum sessions run in parallel for a job: 1.
Maximum elements run in parallel for a session: 1.
Up to 1 processes will be utilized for a job.
Job #1 will run sessions: ,FM0001
cn2107:~ moana004$ --> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR ANTSDIR PYLIBDIR FSLDIR FSLGPUDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder FSL_GPU_SCRIPTS FSLGPUBinary EDDYCUDADIR USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.94.4
User: moana004
System: cn2107
OS: RedHat Linux #1 SMP Tue Jun 28 15:37:28 UTC 2022
------------------------------------------------------------------------
ââââââ\ â ââ\ ââ\
ââ __ââ\ â âââ\ ââ |
ââ / ââ |ââ\ ââ\ â ââââ\ ââ | ââââââ\ ââ\ ââ\
ââ | ââ |ââ | ââ | â ââ ââ\ââ |ââ __ââ\\ââ\ ââ |
ââ | ââ |ââ | ââ | â ââ \ââââ |ââââââââ |\ââââ /
ââ ââ\ââ |ââ | ââ | â ââ |\âââ |ââ ____|ââ ââ\
\ââââââ / \ââââââ | â ââ | \ââ |\âââââââ\ââ /\ââ\
\___âââ\ \______/ â \__| \__| \_______\__/ \__|
\___| â
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSES' which is a part of the QuNex Suite source code package:
https://gitlab.qunex.yale.edu/qunex/qunex/-/tree/master/LICENSES
---> Setting up Octave
........................ Running QuNex v0.94.4 ........................
--- Full QuNex call for command: hcp_pre_freesurfer
gmri hcp_pre_freesurfer --sessionsfolder="/home/moanae/shared/project_FM_Gracely_shared/sessions" --batchfile="/home/moanae/shared/project_FM_Gracely_shared/processing/batch_01_structural.txt" --sessions=",FM0001"
---------------------------------------------------------
WARNING: Parameter qx_cifti_tail was not specified. Its value was imputed from parameter hcp_cifti_tail and set to ''!
WARNING: Parameter qx_nifti_tail was not specified. Its value was imputed from parameter hcp_nifti_tail and set to ''!
WARNING: Parameter cifti_tail was not specified. Its value was imputed from parameter qx_cifti_tail and set to ''!
WARNING: Parameter nifti_tail was not specified. Its value was imputed from parameter qx_nifti_tail and set to ''!
# Generated by QuNex 0.94.4 on 2022-08-09_14.51.44.871388
#
=================================================================
gmri hcp_pre_freesurfer \
--sessionsfolder="/home/moanae/shared/project_FM_Gracely_shared/sessions" \
--sessions="/home/moanae/shared/project_FM_Gracely_shared/processing/batch_01_structural.txt" \
--sessionids=",FM0001" \
=================================================================
Starting multiprocessing sessions in /home/moanae/shared/project_FM_Gracely_shared/processing/batch_01_structural.txt with a pool of 1 concurrent processes
---- Running local
Starting processing of sessions FM0001 at Tuesday, 09. August 2022 14:51:44
Running external command: /opt/HCP/HCPpipelines/PreFreeSurfer/PreFreeSurferPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --t1="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/unprocessed/T1w/FM0001_T1w_MPR1.nii.gz" --t2="NONE" --t1template="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm.nii.gz" --t1templatebrain="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain.nii.gz" --t1template2mm="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm.nii.gz" --t2template="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm.nii.gz" --t2templatebrain="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm_brain.nii.gz" --t2template2mm="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_2mm.nii.gz" --templatemask="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain_mask.nii.gz" --template2mmmask="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm_brain_mask_dil.nii.gz" --brainsize="170" --fnirtconfig="/opt/HCP/HCPpipelines/global/config/T1_2_MNI152_2mm.cnf" --gdcoeffs="NONE" --avgrdcmethod="NONE" --custombrain="NONE" --processing-mode="LegacyStyleData"
You can follow command's progress in:
/home/moanae/shared/project_FM_Gracely_shared/processing/logs/comlogs/tmp_hcp_pre_freesurfer_FM0001_2022-08-09_14.51.44.879913.log
------------------------------------------------------------
------------------------------------------------------------
Session id: FM0001
[started on Tuesday, 09. August 2022 14:51:44]
Running HCP PreFreeSurfer Pipeline [LegacyStyleData] ...
---> T1w image file present.
---> Not using T2w image.
---> WARNING: No distortion correction method specified.
------------------------------------------------------------
Running HCP Pipelines command via QuNex:
/opt/HCP/HCPpipelines/PreFreeSurfer/PreFreeSurferPipeline.sh
--path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp"
--subject="FM0001"
--t1="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/unprocessed/T1w/FM0001_T1w_MPR1.nii.gz"
--t2="NONE"
--t1template="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm.nii.gz"
--t1templatebrain="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain.nii.gz"
--t1template2mm="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm.nii.gz"
--t2template="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm.nii.gz"
--t2templatebrain="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm_brain.nii.gz"
--t2template2mm="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_2mm.nii.gz"
--templatemask="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain_mask.nii.gz"
--template2mmmask="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm_brain_mask_dil.nii.gz"
--brainsize="170"
--fnirtconfig="/opt/HCP/HCPpipelines/global/config/T1_2_MNI152_2mm.cnf"
--gdcoeffs="NONE"
--avgrdcmethod="NONE"
--custombrain="NONE"
--processing-mode="LegacyStyleData"
------------------------------------------------------------
Running HCP PreFS
ERROR: Running HCP PreFS failed with error 1
...
command executed:
/opt/HCP/HCPpipelines/PreFreeSurfer/PreFreeSurferPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --t1="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/unprocessed/T1w/FM0001_T1w_MPR1.nii.gz" --t2="NONE" --t1template="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm.nii.gz" --t1templatebrain="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain.nii.gz" --t1template2mm="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm.nii.gz" --t2template="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm.nii.gz" --t2templatebrain="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm_brain.nii.gz" --t2template2mm="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_2mm.nii.gz" --templatemask="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain_mask.nii.gz" --template2mmmask="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm_brain_mask_dil.nii.gz" --brainsize="170" --fnirtconfig="/opt/HCP/HCPpipelines/global/config/T1_2_MNI152_2mm.cnf" --gdcoeffs="NONE" --avgrdcmethod="NONE" --custombrain="NONE" --processing-mode="LegacyStyleData"
---> logfile: /home/moanae/shared/project_FM_Gracely_shared/processing/logs/comlogs/error_hcp_pre_freesurfer_FM0001_2022-08-09_14.51.44.879913.log
HCP PreFS completed on Tuesday, 09. August 2022 14:51:51
------------------------------------------------------------
===> Final report for command hcp_pre_freesurfer
... FM0001 ---> PreFS failed
===> Not all tasks completed fully!
cn2107:~ moana004$ ls /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/T1w1_gdc.nii.gz
/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/T1w1_gdc.nii.gz
cn2107:~ moana004$ ls -l /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/T1w1_gdc.nii.gz
-r--------. 1 moana004 moanae 7380669 Aug 9 14:23 /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/T1w1_gdc.nii.gz
hcp_pre_freesurfer log file
# Generated by QuNex 0.94.4 on 2022-08-09_14.51.44.879913
#
------------------------------------------------------------
Running external command via QuNex:
/opt/HCP/HCPpipelines/PreFreeSurfer/PreFreeSurferPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --t1="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/unprocessed/T1w/FM0001_T1w_MPR1.nii.gz" --t2="NONE" --t1template="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm.nii.gz" --t1templatebrain="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain.nii.gz" --t1template2mm="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm.nii.gz" --t2template="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm.nii.gz" --t2templatebrain="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm_brain.nii.gz" --t2template2mm="/opt/HCP/HCPpipelines/global/templates/MNI152_T2_2mm.nii.gz" --templatemask="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain_mask.nii.gz" --template2mmmask="/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm_brain_mask_dil.nii.gz" --brainsize="170" --fnirtconfig="/opt/HCP/HCPpipelines/global/config/T1_2_MNI152_2mm.cnf" --gdcoeffs="NONE" --avgrdcmethod="NONE" --custombrain="NONE" --processing-mode="LegacyStyleData"
Test file:
/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/T1w_restore_brain.nii.gz
------------------------------------------------------------
========================================
DIRECTORY: /opt/HCP/HCPpipelines
PRODUCT: HCP Pipeline Scripts
VERSION: v4.4.0-rc-MOD-e7a6af9
COMMIT: e7a6af936d3a9255a2585d2cc16cdea85454e6ca
MODIFIED: YES
========================================
Tue Aug 9 14:51:44 CDT 2022:PreFreeSurferPipeline.sh: HCPPIPEDIR: /opt/HCP/HCPpipelines
Tue Aug 9 14:51:45 CDT 2022:PreFreeSurferPipeline.sh: FSLDIR: /opt/fsl/fsl
Tue Aug 9 14:51:45 CDT 2022:PreFreeSurferPipeline.sh: HCPPIPEDIR_Global: /opt/HCP/HCPpipelines/global/scripts
Tue Aug 9 14:51:45 CDT 2022:PreFreeSurferPipeline.sh: Platform Information Follows:
Linux cn2107 3.10.0-1160.71.1.el7.x86_64 #1 SMP Tue Jun 28 15:37:28 UTC 2022 x86_64 x86_64 x86_64 GNU/Linux
Tue Aug 9 14:51:45 CDT 2022:PreFreeSurferPipeline.sh: Parsing Command Line Options
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: LegacyStyleData mode requested.
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: NOTICE: You are using a mode that enables processing of acquisitions that do not
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: conform to the HCP specification as described in Glasser et al. (2013)!
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: Be aware that if the HCP requirements are not met, the level of data quality
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: can not be guaranteed and the Glasser et al. (2013) paper should not be used
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: in support of this workflow. A manuscript with comprehensive evaluation for
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: the LegacyStyleData processing mode is in active preparation and should be
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: appropriately cited when published.
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: The following LegacyStyleData settings were requested: --t2=NONE
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: Finished Parsing Command Line Options
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: StudyFolder: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: Subject: FM0001
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T1wInputImages: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/unprocessed/T1w/FM0001_T1w_MPR1.nii.gz
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T2wInputImages: NONE
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T1wTemplate: /opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm.nii.gz
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T1wTemplateBrain: /opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain.nii.gz
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T1wTemplate2mm: /opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm.nii.gz
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T2wTemplate: /opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm.nii.gz
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T2wTemplateBrain: /opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm_brain.nii.gz
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T2wTemplate2mm: /opt/HCP/HCPpipelines/global/templates/MNI152_T2_2mm.nii.gz
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: TemplateMask: /opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain_mask.nii.gz
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: Template2mmMask: /opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm_brain_mask_dil.nii.gz
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: BrainSize: 170
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: FNIRTConfig: /opt/HCP/HCPpipelines/global/config/T1_2_MNI152_2mm.cnf
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: MagnitudeInputName:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: PhaseInputName:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: GEB0InputName:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: TE:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: SpinEchoPhaseEncodeNegative:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: SpinEchoPhaseEncodePositive:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: SEEchoSpacing:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: SEUnwarpDir:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T1wSampleSpacing:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T2wSampleSpacing:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: UnwarpDir:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: GradientDistortionCoeffs: NONE
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: AvgrdcSTRING: NONE
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: TopupConfig:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: BiasFieldSmoothingSigma:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: UseJacobian: true
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T1wBiasCorrect:
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: CustomBrain: NONE
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: ProcessingMode: LegacyStyleData
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T1wFolder: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: T2wFolder: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T2w
Tue Aug 9 14:51:48 CDT 2022:PreFreeSurferPipeline.sh: AtlasSpaceFolder: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear
Tue Aug 9 14:51:49 CDT 2022:PreFreeSurferPipeline.sh: POSIXLY_CORRECT=
Tue Aug 9 14:51:49 CDT 2022:PreFreeSurferPipeline.sh: Processing Modality: T1w
Tue Aug 9 14:51:49 CDT 2022:PreFreeSurferPipeline.sh: NOT PERFORMING GRADIENT DISTORTION CORRECTION
[Errno 13] Permission denied: '/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/T1w1_gdc.nii.gz'
Tue Aug 9 14:51:50 CDT 2022:PreFreeSurferPipeline.sh: Not Averaging T1w Images
Tue Aug 9 14:51:50 CDT 2022:PreFreeSurferPipeline.sh: ONLY ONE IMAGE FOUND: COPYING
[Errno 13] Permission denied: '/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/T1w.nii.gz'
Tue Aug 9 14:51:51 CDT 2022:PreFreeSurferPipeline.sh: While running '/opt/HCP/HCPpipelines/PreFreeSurfer/PreFreeSurferPipeline.sh --path=/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp --subject=FM0001 --t1=/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/unprocessed/T1w/FM0001_T1w_MPR1.nii.gz --t2=NONE --t1template=/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm.nii.gz --t1templatebrain=/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain.nii.gz --t1template2mm=/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm.nii.gz --t2template=/opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm.nii.gz --t2templatebrain=/opt/HCP/HCPpipelines/global/templates/MNI152_T2_1mm_brain.nii.gz --t2template2mm=/opt/HCP/HCPpipelines/global/templates/MNI152_T2_2mm.nii.gz --templatemask=/opt/HCP/HCPpipelines/global/templates/MNI152_T1_1mm_brain_mask.nii.gz --template2mmmask=/opt/HCP/HCPpipelines/global/templates/MNI152_T1_2mm_brain_mask_dil.nii.gz --brainsize=170 --fnirtconfig=/opt/HCP/HCPpipelines/global/config/T1_2_MNI152_2mm.cnf --gdcoeffs=NONE --avgrdcmethod=NONE --custombrain=NONE --processing-mode=LegacyStyleData':
Tue Aug 9 14:51:51 CDT 2022:PreFreeSurferPipeline.sh: ERROR: '/opt/fsl/fsl/bin/imcp' command failed with return code: 1
Tue Aug 9 14:51:51 CDT 2022:PreFreeSurferPipeline.sh: ERROR: '/opt/fsl/fsl/bin/imcp' command failed with return code: 1
===> ERROR: Command returned with nonzero exit code
---------------------------------------------------
script: PreFreeSurferPipeline.sh
stopped at line: 570
call: ${RUN} ${FSLDIR}/bin/imcp ${TXwFolder}/${TXwImage}1_gdc ${TXwFolder}/${TXwImage}
expanded call: /opt/fsl/fsl/bin/imcp /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/T1w1_gdc /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/T1w
exit code: 1
---------------------------------------------------
===> Aborting execution!
Hi Estephan,
This now seems like an issue with permission of your file system. I would say that the user in the container has no permissions to work on /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/T1w1_gdc.nii.gz and /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/T1w.nii.gz. Could you check what the permissions for these two files are:
cd /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/T1w/
ls -l
As a test you can try setting all permissions to 777, we do not advocate that you do this for the final processing as you are giving everyone permissions to see your files. But maybe do it as a test so we can see if that is indeed the reason why HCP PreFreeSurfer is unable to access the images. Other than that the logs now look fine!
I changed permissions as suggested and it worked, thank you for your guidance through this process.
cn2107:T1w moana004$ ls -l
total 16896
drwxrwsr-x. 2 moana004 moanae 4096 Aug 9 14:23 ACPCAlignment
-r--------. 1 moana004 moanae 7380669 Aug 9 14:23 T1w1_gdc.nii.gz
-r--------. 1 moana004 moanae 7380669 Aug 9 14:23 T1w.nii.gz
drwxrwsr-x. 2 moana004 moanae 4096 Aug 9 14:23 xfms
cn2107:T1w moana004$ chmod 777 *
cn2107:T1w moana004$ ls -l
total 16896
drwxrwsrwx. 2 moana004 moanae 4096 Aug 9 14:23 ACPCAlignment
-rwxrwxrwx. 1 moana004 moanae 7380669 Aug 9 14:23 T1w1_gdc.nii.gz
-rwxrwxrwx. 1 moana004 moanae 7380669 Aug 9 14:23 T1w.nii.gz
drwxrwsrwx. 2 moana004 moanae 4096 Aug 9 14:23 xfms
Can you tell me who is the user in the container? For future processing, how do you recommend me to proceed? Again, thank you for the help!
So, I assume hcp_pre_freesurfer is now finally running? Well done  . Let me know if some other steps down the line cause you any issues.
. Let me know if some other steps down the line cause you any issues.
In theory, with Singularity you are the same user inside the container. So if you execute stuff through the container it should work. It could be that the 400 privileges that you had set initially were to strict for HCP Pipelines. Like I said 777 is a bit over the top as everyone now has complete access to your data. Try using something more reasonable, e.g. 664. But if this is a piece of HW that only people you trust have access to, then 777 could also be fine.
Yes, hcp_pre_freesurfer finished successfully 
So I never set permissions during all the qunex_container steps, so I do not know why the permissions were set to 400. Should I change anything in the singularity container so the permissions are not an issue for future processing?
I think the permission we are looking at are copied over from the files you are importing. So it seems like you have very strict permission on the original data that is then onboarded?
You’re correct. So I should address the permissions of the raw data files I’m copying in order to process them using qunex. Thanks for the pointer.
Exactly, since users are the ones that need to have full control over who has access to their data, QuNex will not mess around with file permissions. This means that you need to handle the permissions properly outside of QuNex.