Description:
When running the HCP fMRISurface pipeline for legacy data, it did not complete successfully due to missing the T2w_restore.nii.gz - it seems to affect the generation of QC scenes. I did not use T2w during previous steps (structural, fMRIvolume). My questions are: 1. Is this error relevant to fMRISurface processing or can it be ignored? 2. Is there a way to flag the lack of T2w images to fMRISurface when submitting the command?
Thank you.
Call:
cn2107:~ moana004$ qunex_container hcp_fmri_surface --batchfile="${study_sharedfolder}/processing/batch_02_struct-fMRI.txt" --sessionsfolder="${study_sharedfolder}/sessions" --parsessions="1" --parsessions="4" --bolds="auditory|intermingled|mechanical|simultaneous" --bind="${study_sharedfolder}:/${study_sharedfolder}" --container="${HOME}/qunex/qunex_suite-0.94.4.sif"
--> QuNex will run the command over 2 sessions. It will utilize:
Maximum sessions run in parallel for a job: 4.
Maximum elements run in parallel for a session: 1.
Up to 4 processes will be utilized for a job.
Job #1 will run sessions: ,FM0001
cn2107:~ moana004$ --> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR ANTSDIR PYLIBDIR FSLDIR FSLGPUDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder FSL_GPU_SCRIPTS FSLGPUBinary EDDYCUDADIR USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.94.4
User: moana004
System: cn2107
OS: RedHat Linux #1 SMP Tue Jun 28 15:37:28 UTC 2022
------------------------------------------------------------------------
ââââââ\ â ââ\ ââ\
ââ __ââ\ â âââ\ ââ |
ââ / ââ |ââ\ ââ\ â ââââ\ ââ | ââââââ\ ââ\ ââ\
ââ | ââ |ââ | ââ | â ââ ââ\ââ |ââ __ââ\\ââ\ ââ |
ââ | ââ |ââ | ââ | â ââ \ââââ |ââââââââ |\ââââ /
ââ ââ\ââ |ââ | ââ | â ââ |\âââ |ââ ____|ââ ââ\
\ââââââ / \ââââââ | â ââ | \ââ |\âââââââ\ââ /\ââ\
\___âââ\ \______/ â \__| \__| \_______\__/ \__|
\___| â
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSES' which is a part of the QuNex Suite source code package:
https://gitlab.qunex.yale.edu/qunex/qunex/-/tree/master/LICENSES
---> Setting up Octave
........................ Running QuNex v0.94.4 ........................
--- Full QuNex call for command: hcp_fmri_surface
gmri hcp_fmri_surface --sessionsfolder="/home/moanae/shared/project_FM_Gracely_shared/sessions" --parsessions="1" --parsessions="4" --bolds="auditory|intermingled|mechanical|simultaneous" --batchfile="/home/moanae/shared/project_FM_Gracely_shared/processing/batch_02_struct-fMRI.txt" --sessions=",FM0001"
---------------------------------------------------------
WARNING: Parameter qx_cifti_tail was not specified. Its value was imputed from parameter hcp_cifti_tail and set to ''!
WARNING: Parameter qx_nifti_tail was not specified. Its value was imputed from parameter hcp_nifti_tail and set to ''!
WARNING: Parameter cifti_tail was not specified. Its value was imputed from parameter qx_cifti_tail and set to ''!
WARNING: Parameter nifti_tail was not specified. Its value was imputed from parameter qx_nifti_tail and set to ''!
# Generated by QuNex 0.94.4 on 2022-08-11_17.21.00.354162
#
=================================================================
gmri hcp_fmri_surface \
--sessionsfolder="/home/moanae/shared/project_FM_Gracely_shared/sessions" \
--parsessions="4" \
--bolds="auditory|intermingled|mechanical|simultaneous" \
--sessions="/home/moanae/shared/project_FM_Gracely_shared/processing/batch_02_struct-fMRI.txt" \
--sessionids=",FM0001" \
=================================================================
Starting multiprocessing sessions in /home/moanae/shared/project_FM_Gracely_shared/processing/batch_02_struct-fMRI.txt with a pool of 4 concurrent processes
---- Running local
Adding processing of session FM0001 to the pool at Thursday, 11. August 2022 17:21:00
Running external command: /opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_1" --lowresmesh="32" --fmrires="2" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
You can follow command's progress in:
/home/moanae/shared/project_FM_Gracely_shared/processing/logs/comlogs/tmp_hcp_fmri_surface_BOLD_1_FM0001_2022-08-11_17.21.00.389240.log
------------------------------------------------------------
Running external command: /opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_2" --lowresmesh="32" --fmrires="2" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
You can follow command's progress in:
/home/moanae/shared/project_FM_Gracely_shared/processing/logs/comlogs/tmp_hcp_fmri_surface_BOLD_2_FM0001_2022-08-11_17.30.15.122121.log
------------------------------------------------------------
Running external command: /opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_3" --lowresmesh="32" --fmrires="2" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
You can follow command's progress in:
/home/moanae/shared/project_FM_Gracely_shared/processing/logs/comlogs/tmp_hcp_fmri_surface_BOLD_3_FM0001_2022-08-11_17.36.07.622219.log
------------------------------------------------------------
Running external command: /opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_6" --lowresmesh="32" --fmrires="2" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
You can follow command's progress in:
/home/moanae/shared/project_FM_Gracely_shared/processing/logs/comlogs/tmp_hcp_fmri_surface_BOLD_6_FM0001_2022-08-11_17.41.15.765011.log
------------------------------------------------------------
------------------------------------------------------------
Session id: FM0001
[started on Thursday, 11. August 2022 17:21:00]
Running HCP fMRI Surface pipeline [LegacyStyleData] ...
---> PostFS results present.
Skipping the following BOLD images:
... bold4 [rest]
... bold5 [rest]
Running 1 BOLD images in parallel
---> Processing BOLD image 1
... fMRIVolume preprocessed bold image present
------------------------------------------------------------
Running HCP Pipelines command via QuNex:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh
--path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp"
--subject="FM0001"
--fmriname="BOLD_1"
--lowresmesh="32"
--fmrires="2"
--smoothingFWHM="2"
--grayordinatesres="2"
--regname="MSMSulc"
------------------------------------------------------------
Running HCP fMRISurface
---> hcp_fmri_surface test file missing:
/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_1/BOLD_1.dtseries.nii
Try running the command directly for more detailed error information:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_1" --lowresmesh="32" --fmrires="2" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
---> logfile: /home/moanae/shared/project_FM_Gracely_shared/processing/logs/comlogs/error_hcp_fmri_surface_BOLD_1_FM0001_2022-08-11_17.21.00.389240.log
---> Processing BOLD image 2
... fMRIVolume preprocessed bold image present
------------------------------------------------------------
Running HCP Pipelines command via QuNex:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh
--path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp"
--subject="FM0001"
--fmriname="BOLD_2"
--lowresmesh="32"
--fmrires="2"
--smoothingFWHM="2"
--grayordinatesres="2"
--regname="MSMSulc"
------------------------------------------------------------
Running HCP fMRISurface
---> hcp_fmri_surface test file missing:
/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_2/BOLD_2.dtseries.nii
Try running the command directly for more detailed error information:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_2" --lowresmesh="32" --fmrires="2" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
---> logfile: /home/moanae/shared/project_FM_Gracely_shared/processing/logs/comlogs/error_hcp_fmri_surface_BOLD_2_FM0001_2022-08-11_17.30.15.122121.log
---> Processing BOLD image 3
... fMRIVolume preprocessed bold image present
------------------------------------------------------------
Running HCP Pipelines command via QuNex:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh
--path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp"
--subject="FM0001"
--fmriname="BOLD_3"
--lowresmesh="32"
--fmrires="2"
--smoothingFWHM="2"
--grayordinatesres="2"
--regname="MSMSulc"
------------------------------------------------------------
Running HCP fMRISurface
---> hcp_fmri_surface test file missing:
/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_3/BOLD_3.dtseries.nii
Try running the command directly for more detailed error information:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_3" --lowresmesh="32" --fmrires="2" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
---> logfile: /home/moanae/shared/project_FM_Gracely_shared/processing/logs/comlogs/error_hcp_fmri_surface_BOLD_3_FM0001_2022-08-11_17.36.07.622219.log
---> Processing BOLD image 6
... fMRIVolume preprocessed bold image present
------------------------------------------------------------
Running HCP Pipelines command via QuNex:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh
--path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp"
--subject="FM0001"
--fmriname="BOLD_6"
--lowresmesh="32"
--fmrires="2"
--smoothingFWHM="2"
--grayordinatesres="2"
--regname="MSMSulc"
------------------------------------------------------------
Running HCP fMRISurface
---> hcp_fmri_surface test file missing:
/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_6/BOLD_6.dtseries.nii
Try running the command directly for more detailed error information:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_6" --lowresmesh="32" --fmrires="2" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
---> logfile: /home/moanae/shared/project_FM_Gracely_shared/processing/logs/comlogs/error_hcp_fmri_surface_BOLD_6_FM0001_2022-08-11_17.41.15.765011.log
HCP fMRISurface completed on Thursday, 11. August 2022 17:46:00
------------------------------------------------------------
===> Final report for command hcp_fmri_surface
... FM0001 ---> HCP fMRI Surface: bolds 1, 2, 3, 6 failed; 4, 5 skipped
===> Not all tasks completed fully!
Logs:
# Generated by QuNex 0.94.4 on 2022-08-11_17.21.00.389240
#
------------------------------------------------------------
Running external command via QuNex:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_1" --lowresmesh="32" --fmrires="2" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
Test file:
/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_1/BOLD_1.dtseries.nii
------------------------------------------------------------
========================================
DIRECTORY: /opt/HCP/HCPpipelines
PRODUCT: HCP Pipeline Scripts
VERSION: v4.4.0-rc-MOD-e7a6af9
COMMIT: e7a6af936d3a9255a2585d2cc16cdea85454e6ca
MODIFIED: YES
========================================
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: HCPPIPEDIR: /opt/HCP/HCPpipelines
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: FSLDIR: /opt/fsl/fsl
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: CARET7DIR: /opt/workbench/workbench/bin_rh_linux64
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: Platform Information Follows:
Linux cn2107 3.10.0-1160.71.1.el7.x86_64 #1 SMP Tue Jun 28 15:37:28 UTC 2022 x86_64 x86_64 x86_64 GNU/Linux
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: Parsing Command Line Options
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: Path: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: Subject: FM0001
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: NameOffMRI: BOLD_1
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: LowResMesh: 32
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: FinalfMRIResolution: 2
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: SmoothingFWHM: 2
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: GrayordinatesResolution: 2
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: RegName: MSMSulc
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: QCMode: yes
Thu Aug 11 17:21:00 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: RUN:
Thu Aug 11 17:21:01 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: Make fMRI Ribbon
Thu Aug 11 17:21:01 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: mkdir -p /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_1/RibbonVolumeToSurfaceMapping
Thu Aug 11 17:21:01 CDT 2022:RibbonVolumeToSurfaceMapping.sh: HCPPIPEDIR: /opt/HCP/HCPpipelines
Thu Aug 11 17:21:01 CDT 2022:RibbonVolumeToSurfaceMapping.sh: CARET7DIR: /opt/workbench/workbench/bin_rh_linux64
Thu Aug 11 17:21:01 CDT 2022:RibbonVolumeToSurfaceMapping.sh: START
0.507906
0.962613
.7086600
1.2165660
Thu Aug 11 17:26:10 CDT 2022:RibbonVolumeToSurfaceMapping.sh: END
Thu Aug 11 17:26:10 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: Surface Smoothing
Thu Aug 11 17:26:10 CDT 2022:SurfaceSmoothing.sh: HCPPIPEDIR: /opt/HCP/HCPpipelines
Thu Aug 11 17:26:10 CDT 2022:SurfaceSmoothing.sh: CARET7DIR: /opt/workbench/workbench/bin_rh_linux64
Thu Aug 11 17:26:10 CDT 2022:SurfaceSmoothing.sh: START
Thu Aug 11 17:26:27 CDT 2022:SurfaceSmoothing.sh: END
Thu Aug 11 17:26:27 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: Subcortical Processing
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: HCPPIPEDIR: /opt/HCP/HCPpipelines
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: CARET7DIR: /opt/workbench/workbench/bin_rh_linux64
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: START
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: AtlasSpaceFolder: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: ROIFolder: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/ROIs
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: FinalfMRIResolution: 2
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: ResultsFolder: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_1
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: NameOffMRI: BOLD_1
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: SmoothingFWHM: 2
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: BrainOrdinatesResolution: 2
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: VolumefMRI: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_1/BOLD_1
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: Sigma: .84932180028801904272
Thu Aug 11 17:26:27 CDT 2022:SubcorticalProcessing.sh: Creating subject-roi subcortical cifti at same resolution as output
Thu Aug 11 17:26:30 CDT 2022:SubcorticalProcessing.sh: Dilating out zeros
Thu Aug 11 17:26:33 CDT 2022:SubcorticalProcessing.sh: Generate atlas subcortical template cifti
Thu Aug 11 17:26:33 CDT 2022:SubcorticalProcessing.sh: Smoothing and resampling
Thu Aug 11 17:30:01 CDT 2022:SubcorticalProcessing.sh: END
Thu Aug 11 17:30:01 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: Generation of Dense Timeseries
Thu Aug 11 17:30:01 CDT 2022:CreateDenseTimeseries.sh: HCPPIPEDIR: /opt/HCP/HCPpipelines
Thu Aug 11 17:30:01 CDT 2022:CreateDenseTimeseries.sh: CARET7DIR: /opt/workbench/workbench/bin_rh_linux64
Thu Aug 11 17:30:01 CDT 2022:CreateDenseTimeseries.sh: START
Thu Aug 11 17:30:06 CDT 2022:CreateDenseTimeseries.sh: END
Thu Aug 11 17:30:06 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: Generating fMRI QC scene and snapshots
Thu Aug 11 17:30:06 CDT 2022:GenerateFMRIScenes.sh: arguments: --study-folder=/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp --subject=FM0001 --fmriname=BOLD_1 --output-folder=/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_1/fMRIQC
Thu Aug 11 17:30:06 CDT 2022:GenerateFMRIScenes.sh: StudyFolder: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp
Thu Aug 11 17:30:06 CDT 2022:GenerateFMRIScenes.sh: Subject: FM0001
Thu Aug 11 17:30:06 CDT 2022:GenerateFMRIScenes.sh: fMRIName: BOLD_1
Thu Aug 11 17:30:06 CDT 2022:GenerateFMRIScenes.sh: OutputSceneFolder: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_1/fMRIQC
Thu Aug 11 17:30:06 CDT 2022:GenerateFMRIScenes.sh: verboseArg: false
ERRORS loading scene, output image may be incorrect.
NAME OF FILE: T2w_restore.nii.gz
PATH TO FILE: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear
File not found:
ERRORS loading scene, output image may be incorrect.
NAME OF FILE: FM0001.MyelinMap_BC.32k_fs_LR.dscalar.nii
PATH TO FILE: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/fsaverage_LR32k
File not found:
NAME OF FILE: FM0001.SmoothedMyelinMap_BC.32k_fs_LR.dscalar.nii
PATH TO FILE: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/fsaverage_LR32k
File not found:
NAME OF FILE: T2w_restore.nii.gz
PATH TO FILE: /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear
File not found:
Thu Aug 11 17:30:15 CDT 2022:GenerateFMRIScenes.sh: fMRI QC scene generation completed
Thu Aug 11 17:30:15 CDT 2022:GenericfMRISurfaceProcessingPipeline.sh: Completed!
** For an example of how to report an issue, please refer to this post.
Hi, I assume that the FreeSurfer pipeline (pre_FS → FS → post_FS) completed successfully for this subject and also hcp_fmri_volume finished successfully?
Could you please upload the batch file (maybe best as an attachment to your post), so we can see what images your session has? I know you already posted a batch file here, but just to be sure it is the same one.
Yes, all the previous steps completed successfully. I attached here the batch file I used for fMRISurface. FYI, I did small updates to the batch file as I learn the parameters needed for each step.
batch_02_struct-fMRI.txt (42.6 KB)
At a glance all looks good, and this seems like a HCP Pipelines issue. The outgoing call to the script seems OK. Can you provide the contents of:
/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_2/
Also, if possible please upload all log files from structure and volume (pre_fs, fs, post_fs and volume). We also saw that you changed the brain size to 170 mm, what is the rationale here?
Here it is:
cn2107:~ moana004$ ls /home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp/FM0001/MNINonLinear/Results/BOLD_2/
BOLD_2_Atlas.dtseries.nii BOLD_2_SBRef_nomask.nii.gz
BOLD_2_Atlas_mean.dscalar.nii brainmask_fs.2.nii.gz
BOLD_2_Atlas_nonzero.stats.txt fMRIQC
BOLD_2_finalmask.nii.gz Movement_AbsoluteRMS_mean.txt
BOLD_2_finalmask.stats.txt Movement_AbsoluteRMS.txt
BOLD_2_fovmask.nii.gz Movement_Regressors_dt.txt
BOLD_2_Jacobian.nii.gz Movement_Regressors.txt
BOLD_2.L.native.func.gii Movement_RelativeRMS_mean.txt
BOLD_2.nii.gz Movement_RelativeRMS.txt
BOLD_2.R.native.func.gii RibbonVolumeToSurfaceMapping
BOLD_2_SBRef.nii.gz
I attached the logs.
With regards brain size, I followed the suggestion I found here: HCP PreFreeSurfer pipeline — QuNex documentation
–hcp_brainsize … Specifies the size of the brain in mm. 170 is FSL
default and seems to be a good choice, HCP uses
150, which can lead to problems with larger heads
[150].
done_hcp_fmri_volume_BOLD_1_FM0001_2022-08-11_14.06.27.266525.log (70.4 KB)
done_hcp_fmri_volume_BOLD_2_FM0001_2022-08-11_14.50.03.613588.log (36.8 KB)
done_hcp_fmri_volume_BOLD_3_FM0001_2022-08-11_15.27.52.177649.log (36.8 KB)
done_hcp_fmri_volume_BOLD_4_FM0001_2022-08-11_13.50.22.871209.log (67.5 KB)
done_hcp_fmri_volume_BOLD_5_FM0001_2022-08-11_14.24.37.134197.log (29.9 KB)
done_hcp_fmri_volume_BOLD_6_FM0001_2022-08-11_16.07.48.954890.log (36.8 KB)
done_hcp_freesurfer_FM0001_2022-08-10_15.30.01.618021.log (499.2 KB)
done_hcp_post_freesurfer_FM0001_2022-08-11_09.21.20.888751.log (1.8 MB)
done_hcp_pre_freesurfer_FM0001_2022-08-10_14.47.07.894280.log (16.1 KB)
done_import_bids_sub-FM0001_2022-08-09_13.11.53.538376.log (2.6 KB)
I think I know what the error is now. HCP surface uses a hardcoded cift tail value of _Atlas, so you need to set --hcp_cifti_tail="_Atlas". I will set this as the default for the next QuNex version as it makes more sense than to have an empty default. This setting is important for a bunch of HCP steps that you might use later one (e.g. ICAFix, MSMAll).
Let me know if this worked. Thanks!
When trying this solution, I’m having a separate issue. The qunex command is ignoring the “–overwrite” parameter.
cn2107:~ moana004$ qunex_container hcp_fmri_surface --batchfile="${study_sharedfolder}/processing/batch_02_struct-fMRI.txt" --sessionsfolder="${study_sharedfolder}/sessions" --parsessions="1" --parsessions="4" --bolds="rest" --hcp_bold_res="3" --hcp_cifti_tail="_Atlas" --bind="${study_sharedfolder}:/${study_sharedfolder}" --container="${HOME}/qunex/qunex_suite-0.94.4.sif" --overwrite="yes"
--> QuNex will run the command over 2 sessions. It will utilize:
Maximum sessions run in parallel for a job: 4.
Maximum elements run in parallel for a session: 1.
Up to 4 processes will be utilized for a job.
Job #1 will run sessions: ,FM0001
cn2107:~ moana004$ --> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR ANTSDIR PYLIBDIR FSLDIR FSLGPUDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder FSL_GPU_SCRIPTS FSLGPUBinary EDDYCUDADIR USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.94.4
User: moana004
System: cn2107
OS: RedHat Linux #1 SMP Tue Jun 28 15:37:28 UTC 2022
------------------------------------------------------------------------
ââââââ\ â ââ\ ââ\
ââ __ââ\ â âââ\ ââ |
ââ / ââ |ââ\ ââ\ â ââââ\ ââ | ââââââ\ ââ\ ââ\
ââ | ââ |ââ | ââ | â ââ ââ\ââ |ââ __ââ\\ââ\ ââ |
ââ | ââ |ââ | ââ | â ââ \ââââ |ââââââââ |\ââââ /
ââ ââ\ââ |ââ | ââ | â ââ |\âââ |ââ ____|ââ ââ\
\ââââââ / \ââââââ | â ââ | \ââ |\âââââââ\ââ /\ââ\
\___âââ\ \______/ â \__| \__| \_______\__/ \__|
\___| â
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSES' which is a part of the QuNex Suite source code package:
https://gitlab.qunex.yale.edu/qunex/qunex/-/tree/master/LICENSES
---> Setting up Octave
........................ Running QuNex v0.94.4 ........................
--- Full QuNex call for command: hcp_fmri_surface
gmri hcp_fmri_surface --sessionsfolder="/home/moanae/shared/project_FM_Gracely_shared/sessions" --parsessions="1" --parsessions="4" --bolds="rest" --hcp_bold_res="3" --hcp_cifti_tail="_Atlas" --batchfile="/home/moanae/shared/project_FM_Gracely_shared/processing/batch_02_struct-fMRI.txt" --sessions=",FM0001"
---------------------------------------------------------
WARNING: Parameter qx_cifti_tail was not specified. Its value was imputed from parameter hcp_cifti_tail and set to '_Atlas'!
WARNING: Parameter qx_nifti_tail was not specified. Its value was imputed from parameter hcp_nifti_tail and set to ''!
WARNING: Parameter cifti_tail was not specified. Its value was imputed from parameter qx_cifti_tail and set to '_Atlas'!
WARNING: Parameter nifti_tail was not specified. Its value was imputed from parameter qx_nifti_tail and set to ''!
# Generated by QuNex 0.94.4 on 2022-08-12_11.37.21.851524
#
=================================================================
gmri hcp_fmri_surface \
--sessionsfolder="/home/moanae/shared/project_FM_Gracely_shared/sessions" \
--parsessions="4" \
--bolds="rest" \
--hcp_bold_res="3" \
--hcp_cifti_tail="_Atlas" \
--sessions="/home/moanae/shared/project_FM_Gracely_shared/processing/batch_02_struct-fMRI.txt" \
--sessionids=",FM0001" \
=================================================================
Starting multiprocessing sessions in /home/moanae/shared/project_FM_Gracely_shared/processing/batch_02_struct-fMRI.txt with a pool of 4 concurrent processes
---- Running local
Adding processing of session FM0001 to the pool at Friday, 12. August 2022 11:37:21
Running external command: /opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_4" --lowresmesh="32" --fmrires="3" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
Running external command: /opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh --path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp" --subject="FM0001" --fmriname="BOLD_5" --lowresmesh="32" --fmrires="3" --smoothingFWHM="2" --grayordinatesres="2" --regname="MSMSulc"
------------------------------------------------------------
Session id: FM0001
[started on Friday, 12. August 2022 11:37:21]
Running HCP fMRI Surface pipeline [LegacyStyleData] ...
---> PostFS results present.
Skipping the following BOLD images:
... bold1 [auditory]
... bold2 [intermingled]
... bold3 [mechanical]
... bold6 [simultaneous]
Running 1 BOLD images in parallel
---> Processing BOLD image 4
... fMRIVolume preprocessed bold image present
------------------------------------------------------------
Running HCP Pipelines command via QuNex:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh
--path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp"
--subject="FM0001"
--fmriname="BOLD_4"
--lowresmesh="32"
--fmrires="3"
--smoothingFWHM="2"
--grayordinatesres="2"
--regname="MSMSulc"
------------------------------------------------------------
Running HCP fMRISurface --- already completed
---> Processing BOLD image 5
... fMRIVolume preprocessed bold image present
------------------------------------------------------------
Running HCP Pipelines command via QuNex:
/opt/HCP/HCPpipelines/fMRISurface/GenericfMRISurfaceProcessingPipeline.sh
--path="/home/moanae/shared/project_FM_Gracely_shared/sessions/FM0001/hcp"
--subject="FM0001"
--fmriname="BOLD_5"
--lowresmesh="32"
--fmrires="3"
--smoothingFWHM="2"
--grayordinatesres="2"
--regname="MSMSulc"
------------------------------------------------------------
Running HCP fMRISurface --- already completed
HCP fMRISurface completed on Friday, 12. August 2022 11:37:21
------------------------------------------------------------
===> Final report for command hcp_fmri_surface
... FM0001 ---> HCP fMRI Surface: bolds 4, 5 done; 1, 2, 3, 6 skipped
===> Successful completion of all tasks
OK, so the good news is that Surface actually finished and the errors are probably just some weird false alarms raised by HCP Pipelines.
The bad news is that there was an overwrite bug in the qunex_container script. You can get the updated script here qunex_container (40.5 KB). You can also download it through the command from the QuNex quick start.
Got it. The updated qunex_container script now works with the “–overwrite” parameter. Thank you.
Hi,
I also met this problem when I used the qunex hcp_fmri_volume command in a qunex_v0.93.2 container.
I have run other LegacyStyle datasets with this version before and it is stable. When I tried to run the HCPStyle data, something went wrong. Here I prepared to run the fMRI part of HCP EarlyPsy Project which only shared the mini-prepossessing sMRI data. After I set everything up for unprocessed fMRI data and copied T1w and MNINonLinear post-freesurfer data in the directory, I run the command :
qunex hcp_fmri_volume \
--sessions="${study_folder}/${subID}/processing/batch.txt" \
--sessionsfolder="${study_folder}/${subID}/sessions" \
--overwrite="yes"
there seemed to be no error and just skip:
--- Full QuNex call for command: hcp_fmri_volume
gmri hcp_fmri_volume --sessions="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/batch.txt" --sessionsfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions" --overwrite="yes"
---------------------------------------------------------
WARNING: Parameter qx_cifti_tail was not specified. Its value was imputed from parameter hcp_cifti_tail and set to '_Atlas'!
WARNING: Parameter qx_nifti_tail was not specified. Its value was imputed from parameter hcp_nifti_tail and set to ''!
WARNING: Parameter cifti_tail was not specified. Its value was imputed from parameter qx_cifti_tail and set to '_Atlas'!
WARNING: Parameter nifti_tail was not specified. Its value was imputed from parameter qx_nifti_tail and set to ''!
# Generated by QuNex 0.93.2 on 2022-11-15_02.20.56.685175
#
=================================================================
gmri hcp_fmri_volume \
--sessions="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/batch.txt" \
--sessionsfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions" \
--overwrite="yes" \
=================================================================
Starting multiprocessing sessions in /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/batch.txt with a pool of 1 concurrent processes
---- Running local
Starting processing of sessions 1001_01_MR at Tuesday, 15. November 2022 02:20:56
------------------------------------------------------------
Session id: 1001_01_MR
[started on Tuesday, 15. November 2022 02:20:56]
Running HCP fMRI Volume pipeline [HCPStyleData] ...
---> PreFS results present.
---> FS results present.
---> PostFS results present.
---> Looking for spin echo fieldmap set images [PA/AP].
... identified folder SpinEchoFieldMap1
... phase positive PA spin echo fieldmap image present
... phase negative AP spin echo fieldmap image present
... identified folder SpinEchoFieldMap2
... phase positive PA spin echo fieldmap image present
... phase negative AP spin echo fieldmap image present
... TOPUP configuration file present
Skipping the following BOLD images:
... bold1 [REST1]
... bold2 [REST1]
... bold3 [REST2]
... bold4 [REST2]
Running 1 BOLD images in parallel
HCP fMRIVolume completed on Tuesday, 15. November 2022 02:20:56
------------------------------------------------------------
===> Final report for command hcp_fmri_volume
... 1001_01_MR ---> HCP fMRI Volume: bolds 1, 2, 3, 4 skipped
===> Successful completion of all tasks
What should I do to solve this problem?Do I need to upgrade quenx? Other information that may help you to determine the cause of the error is listed below.
- fMRI data import
qunex import_hcp \
--sessionsfolder="${study_folder}/${subID}/sessions" \
--inbox="${data_folder}/${subID}/unprocessed" \
--hcplsname=hcpls
- batch file
Subject_Test_batch.txt (6.8 KB)
- File directory
Yes, please update QuNex a new version (0.95.2) should be up in a couple of hours. The version you are using is quite old. The version specifically addresses some issues with hcp_fmri_volume and BOLD names. I am not sure, but you migh have to rerun setup_hcp for the HCP session file to be recreated properly.
Cheers, Jure
Ok, I’ll wait for the latest version to run. Thanks for your reply and timely help! 
Best, Lianglong
Hi, Jure
I built the qunexcontainer_0.95.2.
singularity build --docker-login qunex_suite-0.95.2.sif docker://gitlab.qunex.yale.edu:5002/qunex/qunexcontainer:0.95.2
But when I used the latest version to run qunex hcp_fmri_volume , I got the same log. It doesn’t work.
........................ Running QuNex v0.95.2 ........................
--- Full QuNex call for command: hcp_fmri_volume
gmri hcp_fmri_volume --sessions="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/batch.txt" --sessionsfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions" --overwrite="yes"
---------------------------------------------------------
WARNING: passing the batchfile through the sessions parameter will be deprecated, please use the batchfile parameter!
WARNING: Parameter qx_cifti_tail was not specified. Its value was imputed from parameter hcp_cifti_tail and set to '_Atlas'!
WARNING: Parameter qx_nifti_tail was not specified. Its value was imputed from parameter hcp_nifti_tail and set to ''!
WARNING: Parameter cifti_tail was not specified. Its value was imputed from parameter qx_cifti_tail and set to '_Atlas'!
WARNING: Parameter nifti_tail was not specified. Its value was imputed from parameter qx_nifti_tail and set to ''!
# Generated by QuNex 0.95.2 on 2022-11-15_20.02.22.350858
#
=================================================================
gmri hcp_fmri_volume \
--sessions="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/batch.txt" \
--sessionsfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions" \
--overwrite="yes" \
=================================================================
Starting multiprocessing sessions in /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/batch.txt with a pool of 1 concurrent processes
---- Running local
Starting processing of sessions 1001_01_MR at Tuesday, 15. November 2022 20:02:22
------------------------------------------------------------
Session id: 1001_01_MR
[started on Tuesday, 15. November 2022 20:02:22]
Running HCP fMRI Volume pipeline [HCPStyleData] ...
---> PreFS results present.
---> FS results present.
---> PostFS results present.
---> Looking for spin echo fieldmap set images [PA/AP].
... identified folder SpinEchoFieldMap1
... phase positive PA spin echo fieldmap image present
... phase negative AP spin echo fieldmap image present
... identified folder SpinEchoFieldMap2
... phase positive PA spin echo fieldmap image present
... phase negative AP spin echo fieldmap image present
... TOPUP configuration file present
Skipping the following BOLD images:
... bold1 [REST1]
... bold2 [REST1]
... bold3 [REST2]
... bold4 [REST2]
Running 1 BOLD images in parallel
HCP fMRIVolume completed on Tuesday, 15. November 2022 20:02:22
------------------------------------------------------------
===> Final report for command hcp_fmri_volume
... 1001_01_MR ---> HCP fMRI Volume: bolds 1, 2, 3, 4 skipped
===> Successful completion of all tasks
Hi!
I am unable to recreate your issue on our end, so I have some additional questions.
I assume that hcp_pre_freesurfer, hcp_freesurfer and hcp_post_freesurfer have been completed for this session?
Also, you ran the setup_hcp and create_batch functions to prepare for HCP processing? Maybe try rerunning these two with the latest version.
Jure
Hi, Jure!
Thanks for your help!
Yes, I think that hcp_pre_freesurfer , hcp_freesurfer and hcp_post_freesurfer have been completed, although the data was downloaded from HCP EarlyPsy Project in NDA.
And I also try to rerun all command using the latest version, but I still got nothing. I rerun that again, and reported all commands and logs as follows:
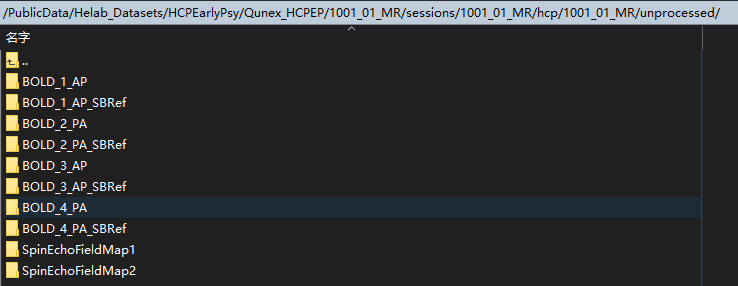
And I attached several pictures about my files and directory.
singularity shell -B /PublicData:/PublicData -B /HeLabData4_master:/HeLabData4_master /HeLabData2/llsun/tool_packages/Container_Package/qunex_suite-0.95.2.sif
Singularity> source /opt/qunex/env/qunex_environment.sh
--> unsetting the following environment variables: PATH MATLABPATH PYTHONPATH QUNEXVer TOOLS QUNEXREPO QUNEXPATH QUNEXLIBRARY QUNEXLIBRARYETC TemplateFolder FSL_FIXDIR FREESURFERDIR FREESURFER_HOME FREESURFER_SCHEDULER FreeSurferSchedulerDIR WORKBENCHDIR DCMNIIDIR DICMNIIDIR MATLABDIR MATLABBINDIR OCTAVEDIR OCTAVEPKGDIR OCTAVEBINDIR RDIR HCPWBDIR AFNIDIR ANTSDIR PYLIBDIR FSLDIR FSLGPUDIR PALMDIR QUNEXMCOMMAND HCPPIPEDIR CARET7DIR GRADUNWARPDIR HCPPIPEDIR_Templates HCPPIPEDIR_Bin HCPPIPEDIR_Config HCPPIPEDIR_PreFS HCPPIPEDIR_FS HCPPIPEDIR_PostFS HCPPIPEDIR_fMRISurf HCPPIPEDIR_fMRIVol HCPPIPEDIR_tfMRI HCPPIPEDIR_dMRI HCPPIPEDIR_dMRITract HCPPIPEDIR_Global HCPPIPEDIR_tfMRIAnalysis HCPCIFTIRWDIR MSMBin HCPPIPEDIR_dMRITractFull HCPPIPEDIR_dMRILegacy AutoPtxFolder FSL_GPU_SCRIPTS FSLGPUBinary EDDYCUDADIR USEOCTAVE QUNEXENV CONDADIR MSMBINDIR MSMCONFIGDIR R_LIBS FSL_FIX_CIFTIRW FSFAST_HOME SUBJECTS_DIR MINC_BIN_DIR MNI_DIR MINC_LIB_DIR MNI_DATAPATH FSF_OUTPUT_FORMAT
Generated by QuNex
------------------------------------------------------------------------
Version: 0.95.2
User: llsun
System: node-3-7
OS: RedHat Linux #83-Ubuntu SMP Wed Jan 18 14:10:15 UTC 2017
------------------------------------------------------------------------
██████\ ║ ██\ ██\
██ __██\ ║ ███\ ██ |
██ / ██ |██\ ██\ ║ ████\ ██ | ██████\ ██\ ██\
██ | ██ |██ | ██ | ║ ██ ██\██ |██ __██\\██\ ██ |
██ | ██ |██ | ██ | ║ ██ \████ |████████ |\████ /
██ ██\██ |██ | ██ | ║ ██ |\███ |██ ____|██ ██\
\██████ / \██████ | ║ ██ | \██ |\███████\██ /\██\
\___███\ \______/ ║ \__| \__| \_______\__/ \__|
\___| ║
DEVELOPED & MAINTAINED BY:
Anticevic Lab, Yale University
Mind & Brain Lab, University of Ljubljana
Murray Lab, Yale University
COPYRIGHT & LICENSE NOTICE:
Use of this software is subject to the terms and conditions defined in
'LICENSES' which is a part of the QuNex Suite source code package:
https://gitlab.qunex.yale.edu/qunex/qunex/-/tree/master/LICENSES
---> Setting up Octave
(/opt/env/qunex) [QuNex ~]$ study_folder=/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP
(/opt/env/qunex) [QuNex ~]$ data_folder=/PublicData/Helab_Datasets/HCPEarlyPsy/imagingcollection01
(/opt/env/qunex) [QuNex ~]$ subID=1001_01_MR
(/opt/env/qunex) [QuNex ~]$ batchfile=/PublicData/Helab_Datasets/HCPEarlyPsy/HCPEP_batch.txt
(/opt/env/qunex) [QuNex ~]$ mkdir ${study_folder}/${subID}
\
--studyfolder="${study_folder}/${subID}"
qunex import_hcp \
--sessionsfolder="${study_folder}/${subID}/sessions" \
--inbox="${data_folder}/${subID}/unprocessed" \
--hcplsname=hcpls
qunex setup_hcp \
--sourcefolder="${study_folder}/${subID}/sessions/${subID}" \
--sourcefile="${study_folder}/${subID}/sessions/${subID}/session_hcp.txt"(/opt/env/qunex) [QuNex ~]$
(/opt/env/qunex) [QuNex ~]$ qunex create_study \
> --studyfolder="${study_folder}/${subID}"
........................ Running QuNex v0.95.2 ........................
--- Full QuNex call for command: create_study
gmri create_study --studyfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR"
---------------------------------------------------------
started running create_study at 2022-11-16 03:39:46, track progress in /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/logs/comlogs/tmp_create_study_2022-11-16_03.39.46.455318.log
call: gmri create_study studyfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR"
-----------------------------------------
Running create_study
===================
Creating study folder structure:
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/analysis
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/analysis/scripts
... folder exists: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing
... folder exists: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/logs
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/logs/batchlogs
... folder exists: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/logs/comlogs
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/logs/runlogs
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/logs/runchecks
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/lists
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/scripts
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/scenes
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/scenes/QC
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/scenes/QC/T1w
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/scenes/QC/T2w
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/scenes/QC/myelin
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/scenes/QC/BOLD
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/scenes/QC/DWI
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/info
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/info/demographics
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/info/tasks
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/info/stimuli
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/info/bids
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/info/hcpls
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/inbox
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/inbox/MR
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/inbox/EEG
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/inbox/BIDS
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/inbox/HCPLS
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/inbox/behavior
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/inbox/concs
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/inbox/events
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/archive
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/archive/MR
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/archive/EEG
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/archive/BIDS
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/archive/HCPLS
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/archive/behavior
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/specs
... created: /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/QC
Preparing template files:
... created parameters_example.txt file
... created .qunexstudy file
-----------------------------------------
Finished at 2022-11-16 03:39:47
===> Successful completion of task
(/opt/env/qunex) [QuNex ~]$
(/opt/env/qunex) [QuNex ~]$ qunex import_hcp \
> --sessionsfolder="${study_folder}/${subID}/sessions" \
> --inbox="${data_folder}/${subID}/unprocessed" \
> --hcplsname=hcpls
........................ Running QuNex v0.95.2 ........................
--- Full QuNex call for command: import_hcp
gmri import_hcp --sessionsfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions" --inbox="/PublicData/Helab_Datasets/HCPEarlyPsy/imagingcollection01/1001_01_MR/unprocessed" --hcplsname="hcpls"
---------------------------------------------------------
started running import_hcp at 2022-11-16 03:39:49, track progress in /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/logs/comlogs/tmp_import_hcp_2022-11-16_03.39.49.056093.log
call: gmri import_hcp sessionsfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions" inbox="/PublicData/Helab_Datasets/HCPEarlyPsy/imagingcollection01/1001_01_MR/unprocessed" hcplsname="hcpls"
-----------------------------------------
Running import_hcp
==================
--> identifying files in /PublicData/Helab_Datasets/HCPEarlyPsy/imagingcollection01/1001_01_MR/unprocessed
--> mapping files to QuNex hcpls folders
--> creating hcpl session 1001_01_MR
--> moving dataset to archive
info: subject 1001, session 01_MR
Running map_hcpls2nii for subject 1001, session 01_MR
=====================================================
--> filesort: name_type_se
--> linked 01.nii.gz <-- 1001_01_MR_SpinEchoFieldMap1_AP.nii.gz
--> linked 02.nii.gz <-- 1001_01_MR_SpinEchoFieldMap1_PA.nii.gz
--> linked 03.nii.gz <-- 1001_01_MR_rfMRI_REST1_AP_SBRef.nii.gz
--> linked 04.nii.gz <-- 1001_01_MR_rfMRI_REST1_AP.nii.gz
--> linked 05.nii.gz <-- 1001_01_MR_rfMRI_REST1_PA_SBRef.nii.gz
--> linked 06.nii.gz <-- 1001_01_MR_rfMRI_REST1_PA.nii.gz
--> linked 07.nii.gz <-- 1001_01_MR_SpinEchoFieldMap2_AP.nii.gz
--> linked 08.nii.gz <-- 1001_01_MR_SpinEchoFieldMap2_PA.nii.gz
--> linked 09.nii.gz <-- 1001_01_MR_rfMRI_REST2_AP_SBRef.nii.gz
--> linked 10.nii.gz <-- 1001_01_MR_rfMRI_REST2_AP.nii.gz
--> linked 11.nii.gz <-- 1001_01_MR_rfMRI_REST2_PA_SBRef.nii.gz
--> linked 12.nii.gz <-- 1001_01_MR_rfMRI_REST2_PA.nii.gz
Final report
============
subject 1001, session 01_MR completed ok. 12 images mapped
-----------------------------------------
Finished at 2022-11-16 03:39:49
===> Successful completion of task
(/opt/env/qunex) [QuNex ~]$
(/opt/env/qunex) [QuNex ~]$ qunex setup_hcp \
> --sourcefolder="${study_folder}/${subID}/sessions/${subID}" \
> --sourcefile="${study_folder}/${subID}/sessions/${subID}/session_hcp.txt"
........................ Running QuNex v0.95.2 ........................
--- Full QuNex call for command: setup_hcp
gmri setup_hcp --sourcefolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/1001_01_MR" --sourcefile="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/1001_01_MR/session_hcp.txt"
---------------------------------------------------------
started running setup_hcp at 2022-11-16 03:39:51, track progress in /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/logs/comlogs/tmp_setup_hcp_2022-11-16_03.39.51.155016.log
call: gmri setup_hcp sourcefolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/1001_01_MR" sourcefile="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/1001_01_MR/session_hcp.txt"
-----------------------------------------
Running setup_hcp
================
===> Setting up HCP folder structure for 1001_01_MR
---> Creating base folder /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions/1001_01_MR/hcp/1001_01_MR/unprocessed
---> creating subfolder BOLD_4_PA
---> linking 12.nii.gz to 1001_01_MR_BOLD_4_PA.nii.gz
---> creating subfolder BOLD_4_PA_SBRef
---> linking 11.nii.gz to 1001_01_MR_BOLD_4_PA_SBRef.nii.gz
---> creating subfolder BOLD_3_AP
---> linking 10.nii.gz to 1001_01_MR_BOLD_3_AP.nii.gz
---> creating subfolder BOLD_3_AP_SBRef
---> linking 09.nii.gz to 1001_01_MR_BOLD_3_AP_SBRef.nii.gz
---> creating subfolder SpinEchoFieldMap2
---> linking 08.nii.gz to 1001_01_MR_BOLD_PA_SB_SE.nii.gz
... SpinEchoFieldMap2 subfolder already exists
---> linking 07.nii.gz to 1001_01_MR_BOLD_AP_SB_SE.nii.gz
---> creating subfolder BOLD_2_PA
---> linking 06.nii.gz to 1001_01_MR_BOLD_2_PA.nii.gz
---> creating subfolder BOLD_2_PA_SBRef
---> linking 05.nii.gz to 1001_01_MR_BOLD_2_PA_SBRef.nii.gz
---> creating subfolder BOLD_1_AP
---> linking 04.nii.gz to 1001_01_MR_BOLD_1_AP.nii.gz
---> creating subfolder BOLD_1_AP_SBRef
---> linking 03.nii.gz to 1001_01_MR_BOLD_1_AP_SBRef.nii.gz
---> creating subfolder SpinEchoFieldMap1
---> linking 02.nii.gz to 1001_01_MR_BOLD_PA_SB_SE.nii.gz
... SpinEchoFieldMap1 subfolder already exists
---> linking 01.nii.gz to 1001_01_MR_BOLD_AP_SB_SE.nii.gz
-----------------------------------------
Finished at 2022-11-16 03:39:51
===> Successful completion of task
(/opt/env/qunex) [QuNex ~]$ cp -r /PublicData/Helab_Datasets/HCPEarlyPsy/fmriresults01/${subID}/MNINonLinear ${study_folder}/${subID}/sessions/${subID}/hcp/${subID}
Data/Helab_Datasets/HCPEarlyPsy/fmriresults01/${subID}/T1w ${study_folder}/${subID}/sessions/${subID}/hcp/${subID}
(/opt/env/qunex) [QuNex ~]$ cp -r /PublicData/Helab_Datasets/HCPEarlyPsy/fmriresults01/${subID}/T1w ${study_folder}/${subID}/sessions/${subID}/hcp/${subID}
(/opt/env/qunex) [QuNex ~]$
(/opt/env/qunex) [QuNex ~]$
(/opt/env/qunex) [QuNex ~]$ qunex create_batch \
> --sessionsfolder="${study_folder}/${subID}/sessions" \
> --paramfile="${batchfile}" \
> --overwrite="yes"
........................ Running QuNex v0.95.2 ........................
--- Full QuNex call for command: create_batch
gmri create_batch --sessionsfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions" --paramfile="/PublicData/Helab_Datasets/HCPEarlyPsy/HCPEP_batch.txt" --overwrite="yes"
---------------------------------------------------------
started running create_batch at 2022-11-16 03:41:07, track progress in /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/logs/comlogs/tmp_create_batch_2022-11-16_03.41.07.428964.log
call: gmri create_batch sessionsfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions" paramfile="/PublicData/Helab_Datasets/HCPEarlyPsy/HCPEP_batch.txt" overwrite="yes"
-----------------------------------------
Running create_batch
====================
---> Creating file batch.txt [/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/batch.txt]
---> appending parameter file [/PublicData/Helab_Datasets/HCPEarlyPsy/HCPEP_batch.txt].
---> Adding: 1001_01_MR
-----------------------------------------
Finished at 2022-11-16 03:41:07
===> Successful completion of task
(/opt/env/qunex) [QuNex ~]$ qunex hcp_fmri_volume \
> --sessions="${study_folder}/${subID}/processing/batch.txt" \
> --sessionsfolder="${study_folder}/${subID}/sessions" \
> --overwrite="yes"
........................ Running QuNex v0.95.2 ........................
--- Full QuNex call for command: hcp_fmri_volume
gmri hcp_fmri_volume --sessions="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/batch.txt" --sessionsfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions" --overwrite="yes"
---------------------------------------------------------
WARNING: passing the batchfile through the sessions parameter will be deprecated, please use the batchfile parameter!
WARNING: Parameter qx_cifti_tail was not specified. Its value was imputed from parameter hcp_cifti_tail and set to '_Atlas'!
WARNING: Parameter qx_nifti_tail was not specified. Its value was imputed from parameter hcp_nifti_tail and set to ''!
WARNING: Parameter cifti_tail was not specified. Its value was imputed from parameter qx_cifti_tail and set to '_Atlas'!
WARNING: Parameter nifti_tail was not specified. Its value was imputed from parameter qx_nifti_tail and set to ''!
# Generated by QuNex 0.95.2 on 2022-11-16_03.41.31.548920
#
=================================================================
gmri hcp_fmri_volume \
--sessions="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/batch.txt" \
--sessionsfolder="/PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/sessions" \
--overwrite="yes" \
=================================================================
Starting multiprocessing sessions in /PublicData/Helab_Datasets/HCPEarlyPsy/Qunex_HCPEP/1001_01_MR/processing/batch.txt with a pool of 1 concurrent processes
---- Running local
Starting processing of sessions 1001_01_MR at Wednesday, 16. November 2022 03:41:31
------------------------------------------------------------
Session id: 1001_01_MR
[started on Wednesday, 16. November 2022 03:41:31]
Running HCP fMRI Volume pipeline [HCPStyleData] ...
---> PreFS results present.
---> FS results present.
---> PostFS results present.
---> Looking for spin echo fieldmap set images [PA/AP].
... identified folder SpinEchoFieldMap1
... phase positive PA spin echo fieldmap image present
... phase negative AP spin echo fieldmap image present
... identified folder SpinEchoFieldMap2
... phase positive PA spin echo fieldmap image present
... phase negative AP spin echo fieldmap image present
... TOPUP configuration file present
Skipping the following BOLD images:
... bold1 [REST1]
... bold2 [REST1]
... bold3 [REST2]
... bold4 [REST2]
Running 1 BOLD images in parallel
HCP fMRIVolume completed on Wednesday, 16. November 2022 03:41:31
------------------------------------------------------------
===> Final report for command hcp_fmri_volume
... 1001_01_MR ---> HCP fMRI Volume: bolds 1, 2, 3, 4 skipped
===> Successful completion of all tasks```



Best, Lianglong
Sorry, the picture are there:
We found that you are using _bolds : rest in the batch file. This means that only BOLDs that have a rest tag will be used (there is no BOLDs in your batch file that are labelled/tagged exactly rest). Please try removing this from the batch file and rerun then.
Thanks for your help again! It worked!
Cheers, Lianglong
Happy to help! Feel free to contact us again if something does not work down the line.