Also, I’m very sorry I have to insert a question about dmri processing in fmri processing issues.
In the following link, I noticed that yjia describes this processing flow, and I processed the dmri data once as described, but the results I got after dwi_parcellate were a bit strange.
[RESOLVED] Dmri processing issues - Use Questions - QuNex Discourse
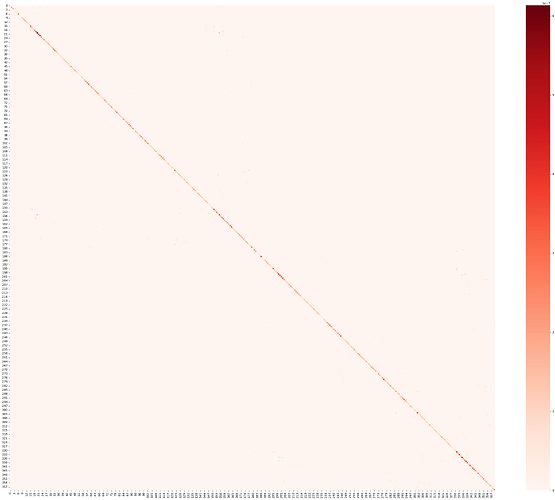
Below is a heatmap of the connection matrix I plotted using waytotnorm_MMP360.pconn.nii obtained by dwi_parcellate, and you can see that its overall value is extremely small (vmax: 6.13359475210018e-07) and the main connections are concentrated in the diagonal region, which is a result that is different from what I was expecting in advance a big difference.
Theoretically shouldn’t the structural connectivity matrix obtained from the above step-by-step procedure be the number of white matter fiber streamlines between any two ROIs? Shouldn’t the resulting structural connectivity matrix be more similar to the common functional connectivity matrix?